Finding genes without DNA
The information produced by population genetic analysis of a breed - in particular, a statistic called "kinship" - can be used to identify groupings of dogs that are more similar to each other genetically than they are to other groups. This technique is called "cluster analysis" and was used in the study of Icelandic Sheepdogs by Oliehoek et al (2009) that we have discussed elsewhere.
The same technique can be used to identify dogs likely to carry the genes for a particular trait (1). A population of 252 Labrador Retrievers owned by the Royal Dutch Guide Dog for the Blind Association were screened for elbow dysplasia using clinical signs of fragmented coronoid process. Seventeen percent of these dogs were found to be affected. Population genetic analysis of the pedigrees of these dogs followed by cluster analysis allowed the researchers to isolate groups of dogs that were more closely related to each other (at least the 0.125 level of relatedness) than to dogs in other groups. They then looked for patterns in the distribution of elbow dysplasia in these genetically distinct clusters.
The same technique can be used to identify dogs likely to carry the genes for a particular trait (1). A population of 252 Labrador Retrievers owned by the Royal Dutch Guide Dog for the Blind Association were screened for elbow dysplasia using clinical signs of fragmented coronoid process. Seventeen percent of these dogs were found to be affected. Population genetic analysis of the pedigrees of these dogs followed by cluster analysis allowed the researchers to isolate groups of dogs that were more closely related to each other (at least the 0.125 level of relatedness) than to dogs in other groups. They then looked for patterns in the distribution of elbow dysplasia in these genetically distinct clusters.
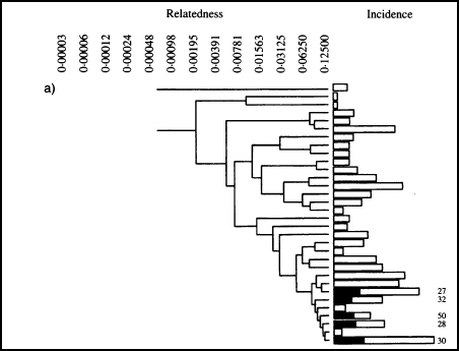
Cluster analysis of Labrador Retrievers in this study. The dogs with affected elbows are indicated by shading.
They found 31 clusters of dogs that had a relatedness at least 0.125, and were genetically distinct from the other clusters. From the dendrogram of the clusters (the funny tree-like graph), it was clear that the cases of elbow dysplasia were all found in five groups of dogs that were closely related to each other. In these 5 clusters, the incidence was greater than 27%, while for all the others it was zero.
What this study shows so clearly is the enormous amount of information that can be leveraged from just pedigrees and some data on occurrence of a particular trait or disease. They were able to accomplish this using relatively small subpopulations of the entire breed, but they note that the most accurate estimations of genetic similarity among individuals would be obtained using pedigrees that extend back to founders.
“A knowledge of the distribution of an inherited disease among clusters of related animals may help to provide the means to select against disease, while maintaining the heterogeneity of the population. The clusters of dogs at risk may also provide a basis for the selection of animals for molecular genetic studies to search for markers of the genes underlying the disease.”
1) Ubbink, GJ, HAW Hazewinkel, J Rothuizen, J van de Broek, & WTC Wolvekamp. 1998. Cluster analysis of the genetic heterogeneity and disease distributions in purebred dog populations. Veterinary Record 142: 209-213.
What this study shows so clearly is the enormous amount of information that can be leveraged from just pedigrees and some data on occurrence of a particular trait or disease. They were able to accomplish this using relatively small subpopulations of the entire breed, but they note that the most accurate estimations of genetic similarity among individuals would be obtained using pedigrees that extend back to founders.
“A knowledge of the distribution of an inherited disease among clusters of related animals may help to provide the means to select against disease, while maintaining the heterogeneity of the population. The clusters of dogs at risk may also provide a basis for the selection of animals for molecular genetic studies to search for markers of the genes underlying the disease.”
1) Ubbink, GJ, HAW Hazewinkel, J Rothuizen, J van de Broek, & WTC Wolvekamp. 1998. Cluster analysis of the genetic heterogeneity and disease distributions in purebred dog populations. Veterinary Record 142: 209-213.

