THE IMPORTANCE OF HISTORY
The Pedigree Database
Your genetic makeup is the result of your genealogical history - who your parents were, who their parents were, the "grand" and "great-grand" and "great-great-grand" ancestors - all contributed something to the genes on your chromosomes. The same of course is true for dogs, and for recognized breeds there are pedigree databases that document ancestry. These records can be incomplete and they also usually contain errors, but they do provide a place to start if you want to understand the history behind the genetics of a particular dog.
When you breed dogs, selecting a particular set of parents and evaluating offspring for merit, you are evaluating phenotype - the observable traits of the dog, which can include structure, behavior, intelligence, and other breed traits, as well as diseases, disorders, and other triats that are not always evident. Of course, what you're really trying to do as a breeder is put the right combination of genes in a dog to produce the qualities that you want. For today's dog breeders, phenotype is a proxy for genetic selection.
But if you knew more about the genotype (genetic composition) of your dogs, your breeding choices could be informed not just by phenotype but also by good predictions of the genetic makeup of a particular animal. This would be more efficient than using only phenotype, and it doesn't even require DNA analysis. All we need to know is history.
The pedigree of a dog is documentation of its own genetic history as well as its relationships with all the other members of its breed. Since we know that every animal gets half of its genes from the sire and half from the dam, there is some predictability in genetic inheritance. We can't predict which gene an animal will inherit from each parent, but we do know that it will always be (exactly!) half. With a little math and probability theory, we can go from assessing piles of paper (or digital) pedigrees to statistical predictions of the genetics behind the phenotype of a particular dog. Instead of trying to guess whether a sire will produce offspring with good hips or bad based, we can actually assign a number to the "genetic value" of a dog that tells us something about the quality of the hips in his offspring. In fact, the sire himself might have poor hip scores, yet have a high genetic value for hips, which means that an animal that might have been eliminated from the breeding pool instead contributes to improving the hips in the breed. We don't need to know anything about the genes or inheritance involved in hip dysplasia. We can do this from a pedigree database and information about the phenotype of the dog's close relatives.
A good pedigree database can provide extremely useful information to breeders, and the work involved in putting one together and maintaining it will be rewarded by the information it will provide that you can't access any other way. A primary goal of every ICB Breeding for the Future breed group is to get that pedigree database together so that we can begin to use it as a basic tool to inform breeding decisions and selection based on genotype and not just phenotype.
Your genetic makeup is the result of your genealogical history - who your parents were, who their parents were, the "grand" and "great-grand" and "great-great-grand" ancestors - all contributed something to the genes on your chromosomes. The same of course is true for dogs, and for recognized breeds there are pedigree databases that document ancestry. These records can be incomplete and they also usually contain errors, but they do provide a place to start if you want to understand the history behind the genetics of a particular dog.
When you breed dogs, selecting a particular set of parents and evaluating offspring for merit, you are evaluating phenotype - the observable traits of the dog, which can include structure, behavior, intelligence, and other breed traits, as well as diseases, disorders, and other triats that are not always evident. Of course, what you're really trying to do as a breeder is put the right combination of genes in a dog to produce the qualities that you want. For today's dog breeders, phenotype is a proxy for genetic selection.
But if you knew more about the genotype (genetic composition) of your dogs, your breeding choices could be informed not just by phenotype but also by good predictions of the genetic makeup of a particular animal. This would be more efficient than using only phenotype, and it doesn't even require DNA analysis. All we need to know is history.
The pedigree of a dog is documentation of its own genetic history as well as its relationships with all the other members of its breed. Since we know that every animal gets half of its genes from the sire and half from the dam, there is some predictability in genetic inheritance. We can't predict which gene an animal will inherit from each parent, but we do know that it will always be (exactly!) half. With a little math and probability theory, we can go from assessing piles of paper (or digital) pedigrees to statistical predictions of the genetics behind the phenotype of a particular dog. Instead of trying to guess whether a sire will produce offspring with good hips or bad based, we can actually assign a number to the "genetic value" of a dog that tells us something about the quality of the hips in his offspring. In fact, the sire himself might have poor hip scores, yet have a high genetic value for hips, which means that an animal that might have been eliminated from the breeding pool instead contributes to improving the hips in the breed. We don't need to know anything about the genes or inheritance involved in hip dysplasia. We can do this from a pedigree database and information about the phenotype of the dog's close relatives.
A good pedigree database can provide extremely useful information to breeders, and the work involved in putting one together and maintaining it will be rewarded by the information it will provide that you can't access any other way. A primary goal of every ICB Breeding for the Future breed group is to get that pedigree database together so that we can begin to use it as a basic tool to inform breeding decisions and selection based on genotype and not just phenotype.
Reconstructing Genetic History
The genetic status of a breed today is the result of its history, and we can learn much about that from the pedigree records. Under the "Breed Projects" tab on the ICB website, you will find ongoing analysis of several breeds we are working on right now. Let's have a look at a few of these for examples of what we can learn.
The Chinook
The beginning of the breed
The beginning of the Chinook is fairly well documented and you can review the details of the history elsewhere. For our purposes, we want to identify the dogs who contributed to the gene pool of what eventually became the Chinook breed.
The founding sire was named Chinook, and he was the progeny of a mastiff-type dog (some sources say St Bernard) named Kim and a dog named Ningo who was the granddaughter of a Greenland Dog named Polaris. We don't know anything about the generation between Polaris and Ningo, so we have no way to know the genetic composition of Ningo, but we would expect about 12% of Chinook's genome to originate from Polaris. We know that 50% of Chinook's genes are from
So Chinook would appear to be a mix that included some molossor and spitz blood.
Chinook was bred to a German Shepherd and a Belgian Shepherd, named Erika and Myrra and respectively. One puppy from the mating with Erika appears in the pedigree history, a bitch named Laska; Myrra produced two puppies, Jock and Zembla.
So starting with three unrelated dogs (Chinook, Myrra, and Erika), we 50% of Erika's genome because she produced only one male puppy (Hootchinoo, sired by Chinook) that produced offspring of its own; in the first generation therefore we go from 3 dogs to 2.5 dogs "genetically". Further, because her puppy was male, her mtDNA is lost to the breed at this generation. From this, we known then that the mtDNA in all Chinooks must have originated from Myrra (the Belgian Shepherd).
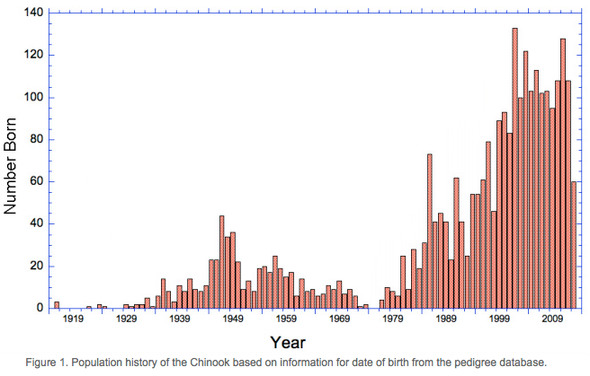
Variations in population size
The genetic status of a breed today is the result of its history, and we can learn much about that from the pedigree records. Under the "Breed Projects" tab on the ICB website, you will find ongoing analysis of several breeds we are working on right now. Let's have a look at a few of these for examples of what we can learn.
The Chinook
The beginning of the breed
The beginning of the Chinook is fairly well documented and you can review the details of the history elsewhere. For our purposes, we want to identify the dogs who contributed to the gene pool of what eventually became the Chinook breed.
The founding sire was named Chinook, and he was the progeny of a mastiff-type dog (some sources say St Bernard) named Kim and a dog named Ningo who was the granddaughter of a Greenland Dog named Polaris. We don't know anything about the generation between Polaris and Ningo, so we have no way to know the genetic composition of Ningo, but we would expect about 12% of Chinook's genome to originate from Polaris. We know that 50% of Chinook's genes are from
So Chinook would appear to be a mix that included some molossor and spitz blood.
Chinook was bred to a German Shepherd and a Belgian Shepherd, named Erika and Myrra and respectively. One puppy from the mating with Erika appears in the pedigree history, a bitch named Laska; Myrra produced two puppies, Jock and Zembla.
So starting with three unrelated dogs (Chinook, Myrra, and Erika), we 50% of Erika's genome because she produced only one male puppy (Hootchinoo, sired by Chinook) that produced offspring of its own; in the first generation therefore we go from 3 dogs to 2.5 dogs "genetically". Further, because her puppy was male, her mtDNA is lost to the breed at this generation. From this, we known then that the mtDNA in all Chinooks must have originated from Myrra (the Belgian Shepherd).
Variations in population size