Why have we made so little progress in 50 years? I think there are a number of issues, but one of the most important is that we don't worry as much as we could about environmental (i.e., non-genetic) factors.
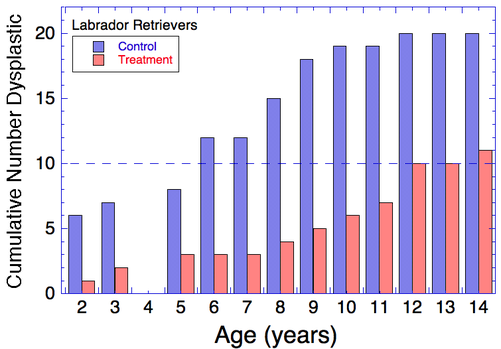
| For example, these are data for Labrador Retrievers raised from birth under common conditions in a research facility. From each litter of puppies, one puppy of a pair went into the control group and the sibling went into the treatment group. The dogs were raised through adulthood and every year (but one) their hips were evaluated for evidence of dysplasia. The only difference between the control and treatment groups was how much they were fed. The results of this simple experiment were astonishing. In Labradors that were fed the normal amount of food, more than half had evidence of hip dysplasia by 6 years of age and most by about 12 years. In the treatment group that was fed less, half of the dogs were still free of dysplasia at 12 years old. |
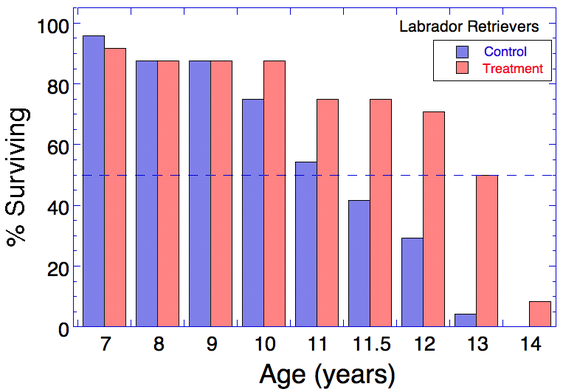
The food restricted group not only suffered less pain and loss of mobility, they also lived longer. Only about half of the control group lived longer than 11 years and only 30% survived to 12 years. But about 75% of the dogs in the treatment group survived to 11 years and 50% lived to 13.
Think about this. We can make a dramatic difference in the risk of hip dysplasia and quality of life of a dog just by adjusting food intake. If I sold a pill that would make a huge difference like this, I'd be rich and your dog would be much happier.
You have probably heard of many things that are claimed to prevent hip dysplasia. Vitamin C, raw diet, mineral supplements, specific forms of exercise, and on and on. But most of these failed to prove beneficial in careful scientific experiments. Don't waste your time and money on supposed treatments that make no difference. Focus on the things that have been proven to matter and that you can control.
Food just one of the many non-genetic factors that can affect the development of hip dysplasia in dogs. There is much more we can do that will reduce risk and make a real difference in quality of life after the breeding decision is made. You can learn more about the cause and prevention of hip and elbow dysplasia in dog in ICB's online course.
|
Learn more about how genes and environment affect the risk of hip and elbow dysplasia, as well as how to use heritability and Estimated Breeding Values (EBVs) to dramatically improve the efficiency of genetic selection. The 10 week course is online and you can work at your own pace! |
Smith, GK, ER Paster, MY Powers, DF Lawler, DN Biery, FS Shofer, PJ McKellvie & RD Kealy. 2006. Lifelong diet restriction and radiographic evidence of osteoarthritis of the hip joint in dogs. JAVMA 229: 690-693.
ICB's online courses
***************************************
Visit our Facebook Groups
ICB Institute of Canine Biology
...the latest canine news and research
ICB Breeding for the Future
...the science of animal breeding