- Bellumori TP, TR Famula, DL Bannasch, JM Belanger, & AM Oberbauer 2013 Prevalence of inherited disorders among mixed-breed and purebred dogs: 27,254 cases (1995-2010). J Am Vet Med Assoc 242: 1549-1555. (pdf)
NOTE: The data from which the information presented here was derived is appended at the bottom.
What makes this complicated is that there are many purebred dogs that live long, happy lives, and others that do suffer from an inherited disorder, and of course the same can be said of the mutt of mixed heritage. To get beyond the anecdotes, what we need are data, and a recent study provides them.
The study by Bellumori et al (2013) used medical records from the veterinary clinic at UC Davis for more than 27,000 dogs and compared the incidence of 24 genetic disorders in mixed versus purebred dogs. The abstract of the paper is included at the bottom of this page.
Here is what they found:
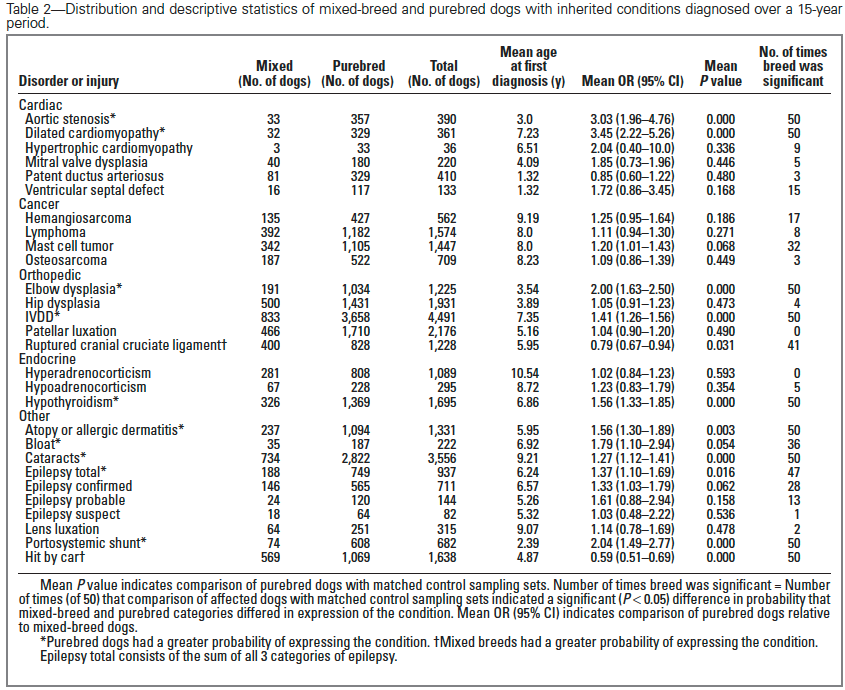
1) The incidence of 10 genetic disorders (42%) was significantly greater in purebred dogs.
2) The incidence of 1 disorder (ruptured cranial cruciate ligament; 4%) was greater in mixed breed dogs.
3) For the rest of the disorders examined, they found no difference in incidence between mixed and purebred dogs.
| MORE IN PUREBREEDS Aortic stenosis Dilated cardiomyopathy Elbow dysplasia IVDD Hypoadrenocorticism Atopy / allergic dermatitis Bloat Cataracts Epilepsy (total) Portosystemic shunt | MORE IN MIXED BREEDS Ruptured cranial cruciate ligament | NO DIFFERENCE Hypertrophic cardiomyopathy Mitral valve dysplasia Patent ductus arteriosus Ventricular septal defect Hemangiosarcoma Lymphoma Mast cell tumor Osteosarcoma Hip dysplasia Patellar luxation Hyperadrenocortism Hypothyroidism Lens luxation Epilepsy (confirmed) Epilepsy (probable) Epilepsy (suspect) |
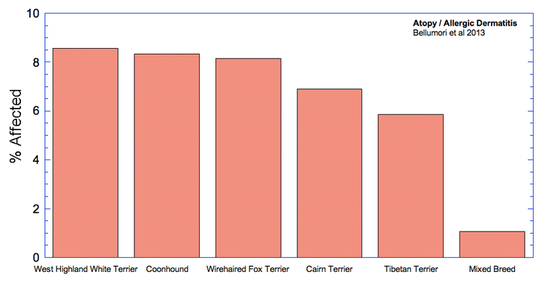
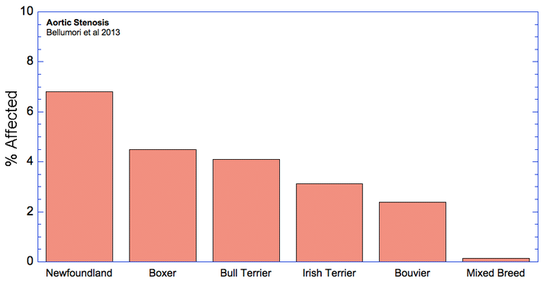
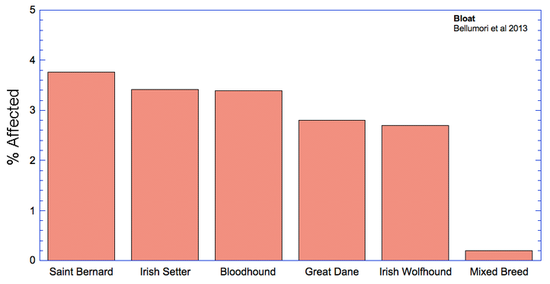
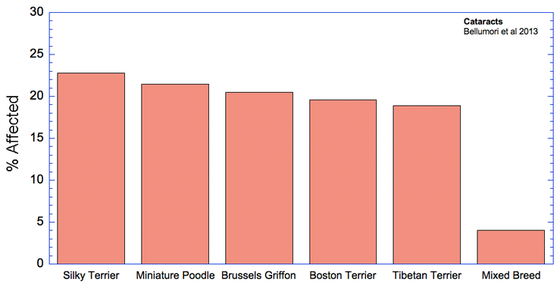
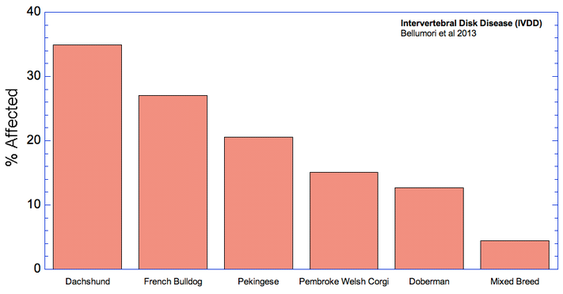
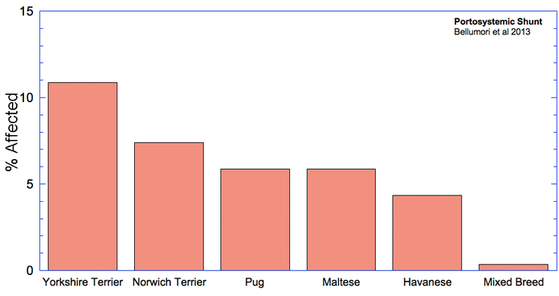
The data in the study are presented as a mean odds ratio (OR) comparing purebred to mixed breed dogs. An equal risk for a disorder in purebred and mixed breed dogs would have an OR = 1. To make the data easier to visualize, I have computed (OR* = 1-OR), for which equal risk in purebred and mixed breeds dogs would have a value of 0, and higher odds for purebreds would have OR* > 0, and higher odds for mixed breeds would have OR* < 0. Therefore, in the graph below, positive values (above the horizontal line) indicate that the odds risk is greater for purebreds, and negative values indicate greater risk for mixed breeds. Asterisks (*) on bars indicate that in 50 comparisons of affected dogs with matched control sampling sets, there was a significant probability (p < 0.05; i.e., less than 5%) that pure and mixed breed dogs differed in expression of the condition. (See Methods and Materials: Statistical Analysis in Bellumori et al 2013 for details.)
An interesting thing to note is that with the exception of one disorder, patent ductus arterioles, the odds ratios are higher for purebreds, but these failed to meet the criterion for statistical significance (hence no asterisk). This does not mean that the incidence in purebreds and mixed breeds was the same, only that they failed to find a statically significant difference in this study. This might be because they had inadequate data to detect a difference, or that in fact purebred and mixed breed dogs are the same and the differences in these data can simply be an artifact of sampling. (They incidentally found a significantly higher risk of being hit by a car in mixed breed dogs, which of course is not a health disorder and presumably not genetic.)
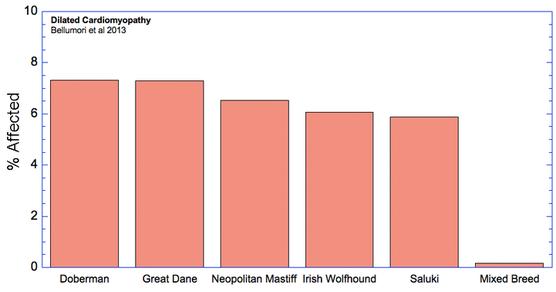
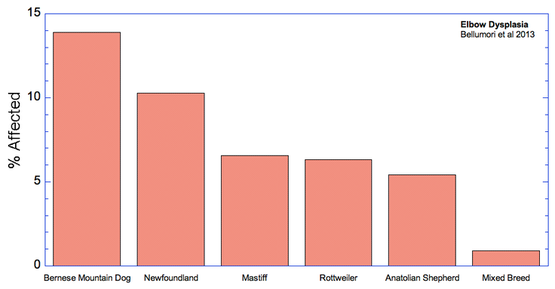
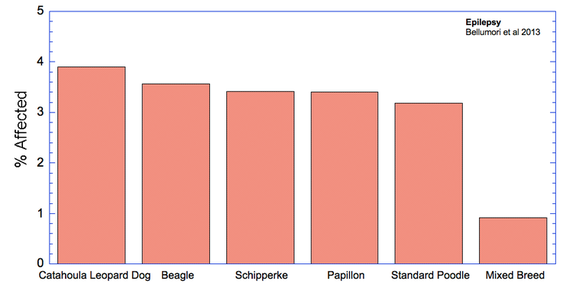
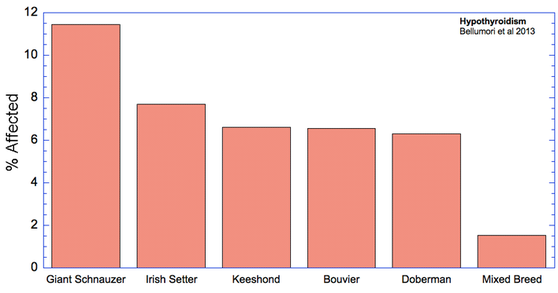
For the 10 genetic disorders which were significantly more prevalent in purebred dogs, I have created graphs that include the 5 breeds with the highest rates of affected dogs plus the value for mixed breeds.
Nevertheless, the findings of this study in the lay press and among purebred fanciers were not so clearly expressed. Some examples:
- "It has been publicly discussed for years that hereditary disorders would be a direct consequence of the strict selective breeding of pedigree dogs and that for this reason the purebreds would have a much greater risk of developing hereditary disorders than mixed breed dogs. According to the latest research by Bellumori and his group, this assumption does not seem to hold. Indeed many diseases seem to be as common in mixed breed as in pedigree dogs" (Moller, on the MyDogDNA website; pdf)
- "A new study on the prevalence of inherited disorders among American mixed breed and purebred dogs has negated the common assumption that a mixed breed dog is always healthier than a purebred dog" (Quickfall 2013).
- "A new study by researchers at the University of California, Davis, indicates that mixed breeds don’t necessarily have an advantage when it comes to inherited canine disorders." UC Davis press release
And another hedge: "mixed breeds don't necessarily (emphasis mine) have an advantage when it comes to inherited canine disorders." This would seem to be saying that the default argument is that mixed breeds are healthier, usually but not necessarily, whatever that means. So indeed, "this assumption does not seem to hold", but for nearly half the disorders examined here the mixed breeds came out on top.
This study found that purebred dogs have a significantly greater risk of developing many of the hereditary disorders examined in this study. No, mixed breed dogs are not ALWAYS healthier than purebreds; and also, purebreds are not "as healthy" as mixed breed dogs. The results of this study will surprise nobody who understands the basics of Mendelian inheritance. Breeding related animals increases the expression of genetic disorders caused by recessive mutations, and it also increases the probability of producing offspring that will inherit the assortment of genes responsible for a polygenic disorder.
The authors of this study tackled a very important question that is difficult to address because collecting the "perfect" data set is impossible. Using data on clinical occurrence of disease is fraught with difficulty because of many sources of potential complication - perhaps purebred dogs are more likely to receive veterinary treatment than mixed breeds, and comparisons among groups (e.g., afflicted vs not, purebred vs mixed) are confounded by unequal sample sizes or differences among groups in the age, sex, etc of animals. It's a statistician's nightmare. (In fact, a highly regarded statistician, Thomas Famula, was involved in the study.) In fact, the "perfect" comparison will never be done. But this study presents a large compilation of data and a thorough analysis that is the first (and might be the only) attempt to explore differences in predisposition to disease in purebred and mixed breed dogs.
- Moller F Mixed breed dogs are not protected from breed disease heritage. MyDogDNA website. (pdf)
- Quickfall L 2013 Kennel Club welcomes study looking at health of all dogs. Dog News, Vol 29(30): 134, July 26, 2013. http://issuu.com/dognews/docs/072613/134
- UC Davis press release (4/2/2014) Purebred dogs not always at higher risk for genetic disorders, study finds. (pdf)
- Wood R 2013 Prevalence of genetic disorders compared in purebred and mixed-breed dogs. CABI VetMed Resource. http://www.cabi.org/VetMedResource/news/23088