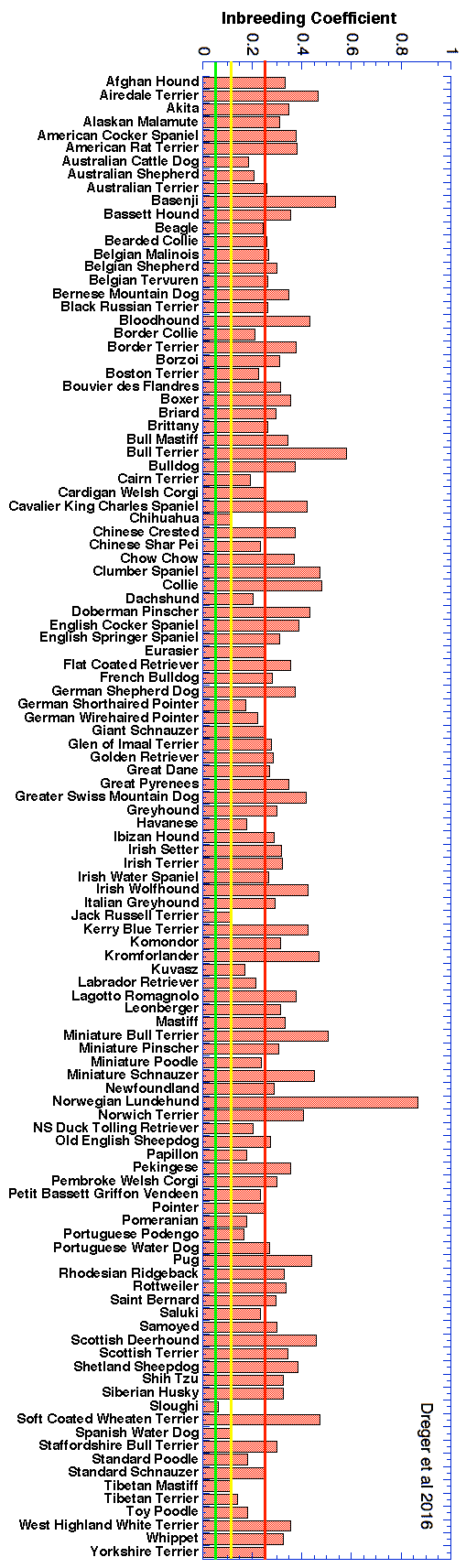
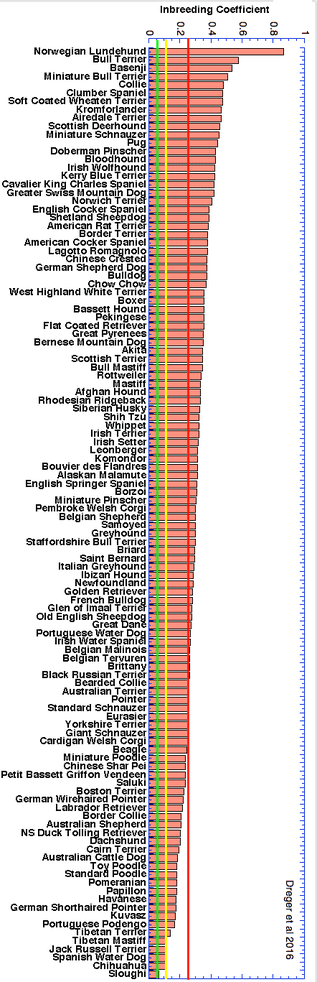
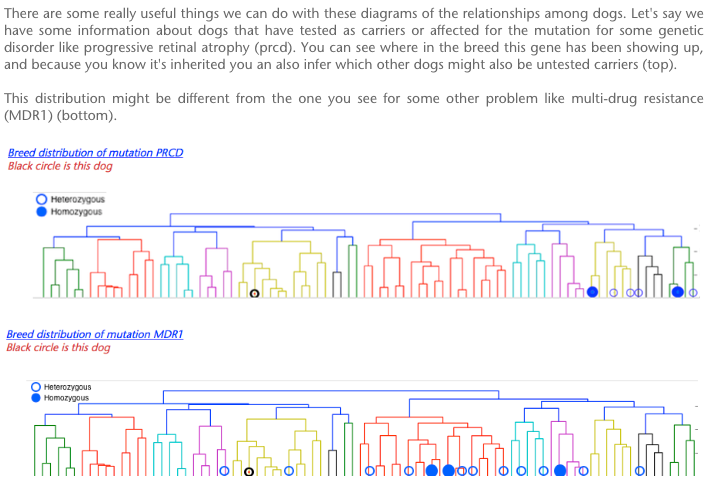
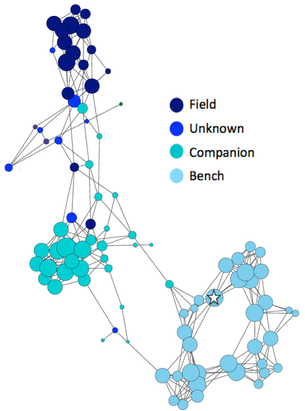
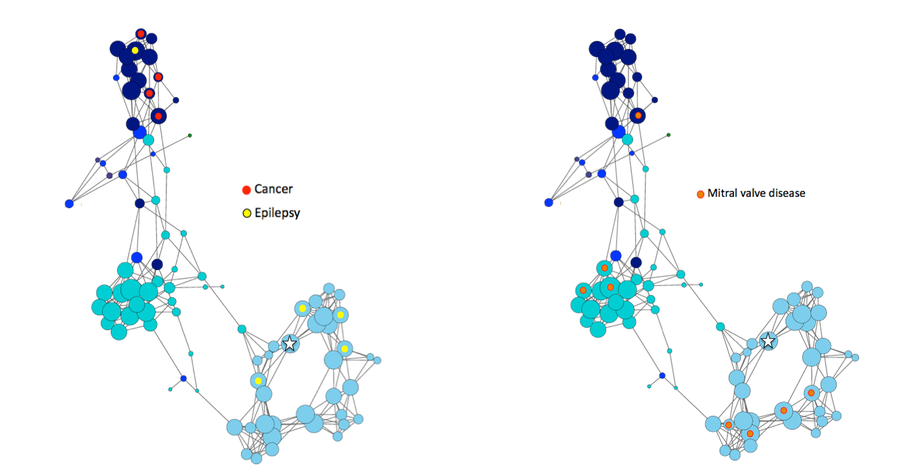
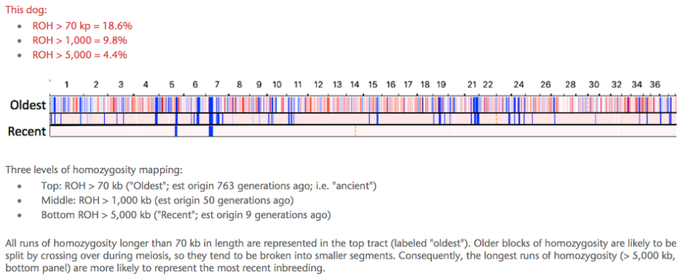
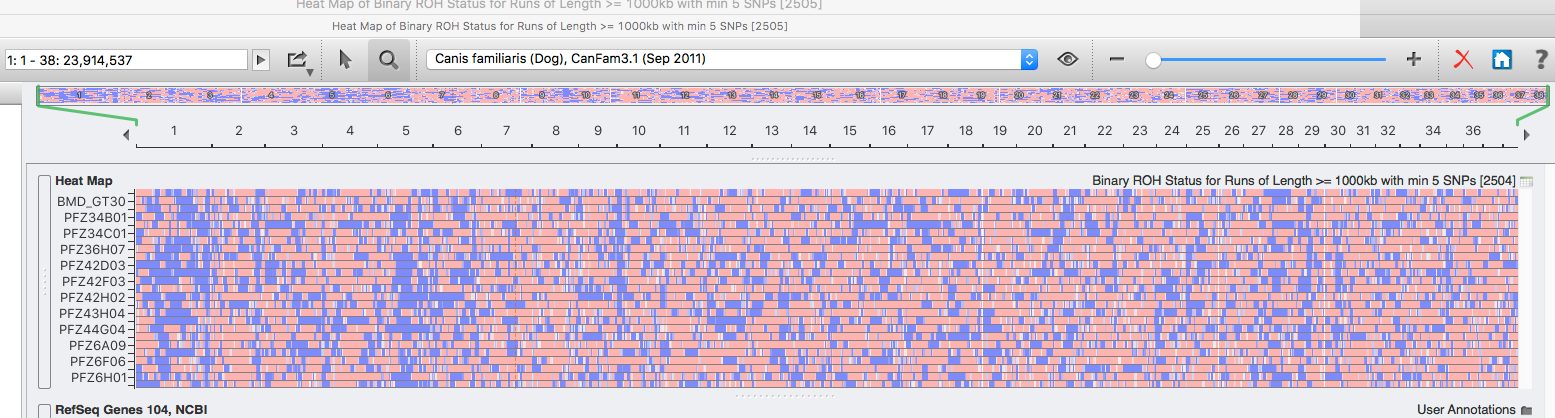
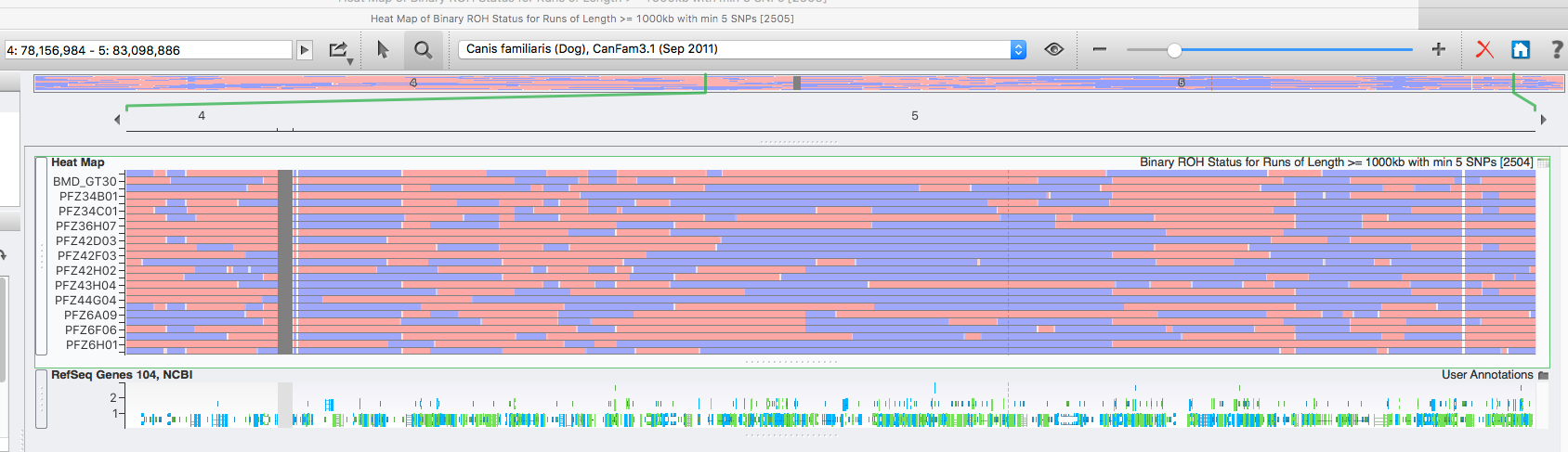
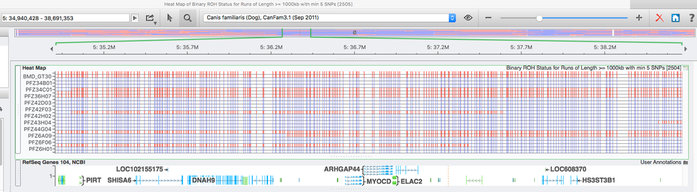
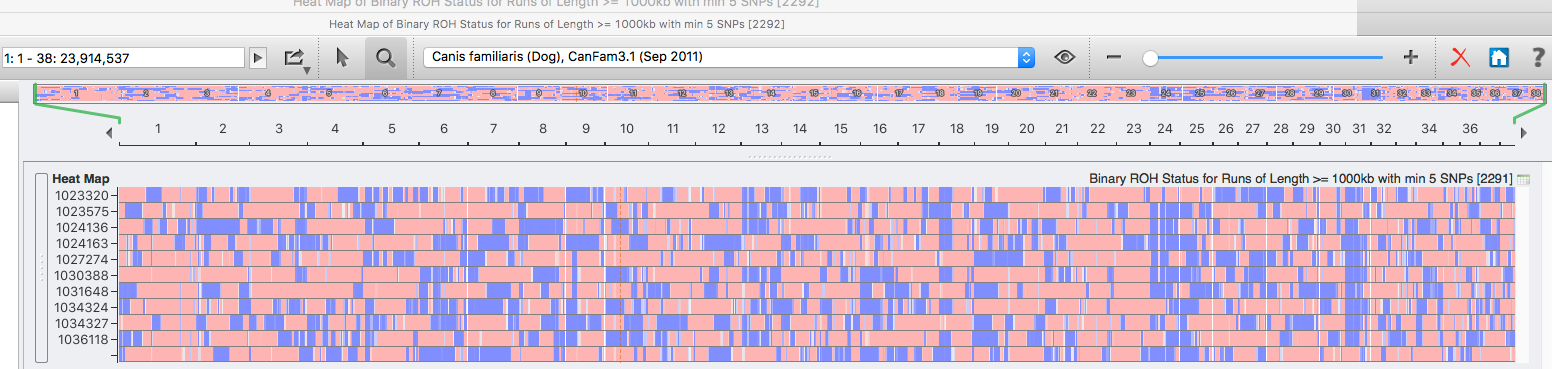
The highest level of inbreeding by far (> 80%) is for the Norwegian Lundehund. This breed suffers from extremely low fertility and high puppy mortality as well as an often lethal gastrointestinal disorder. As a last effort to save it from extinction, it is now the focus of a cross-breeding program supported by the Norwegian Lundehund club and guided by an international group of scientists who are now reporting the successful production of F2 offspring.
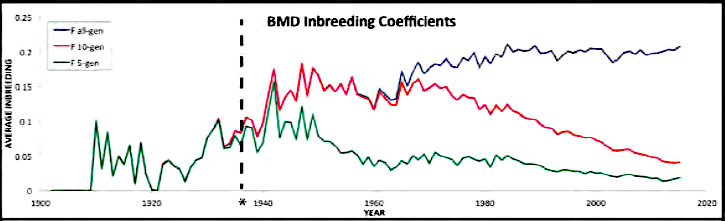
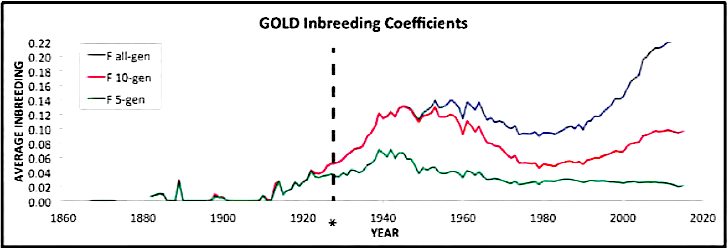
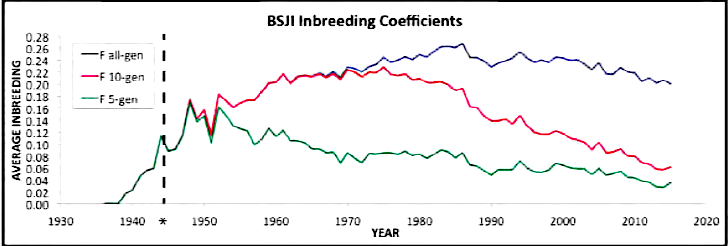
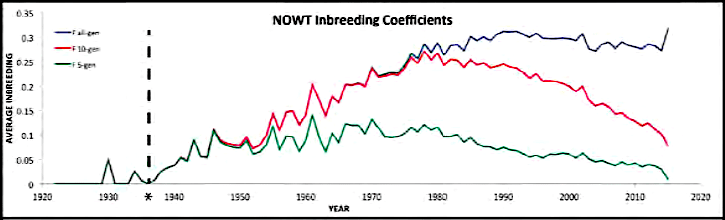
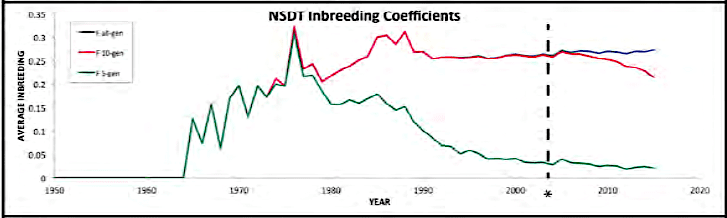
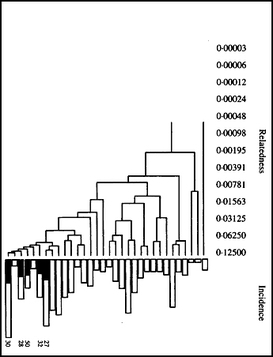
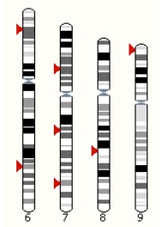
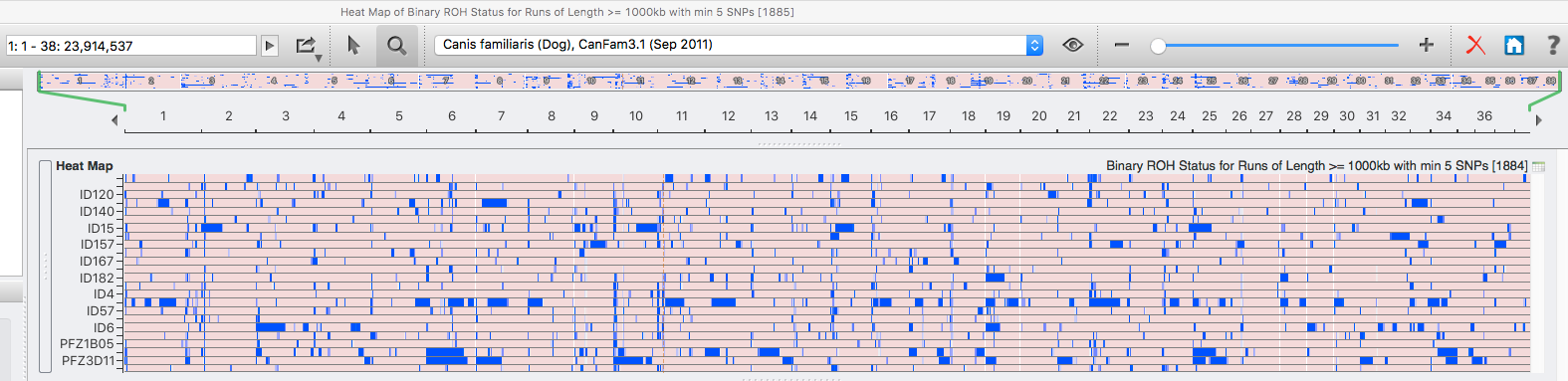
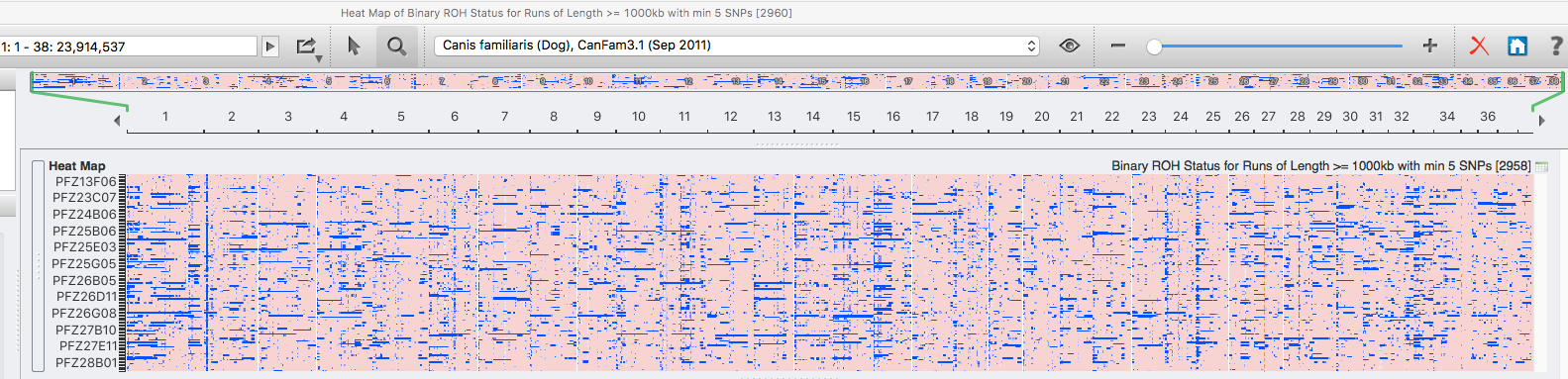
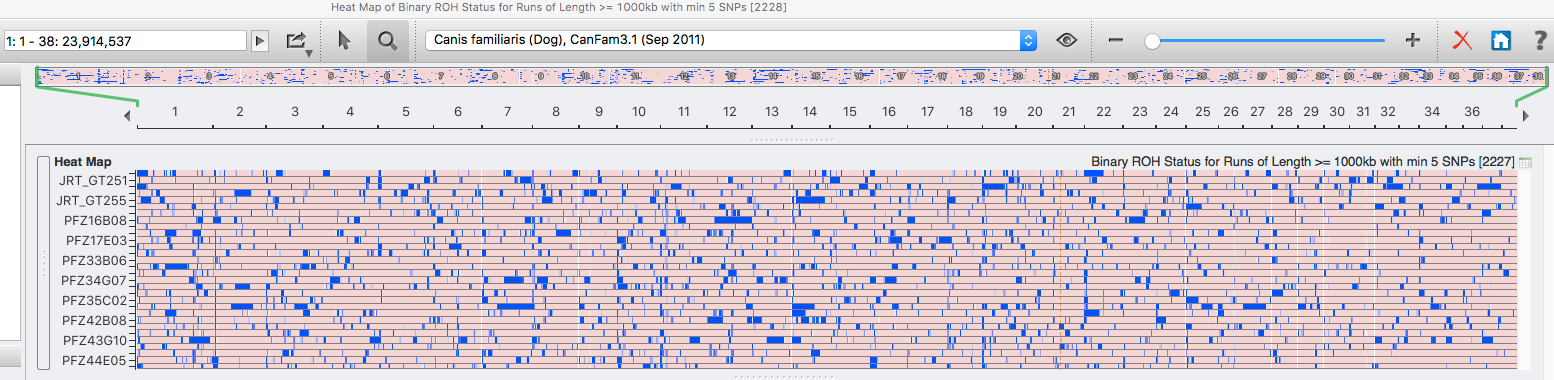
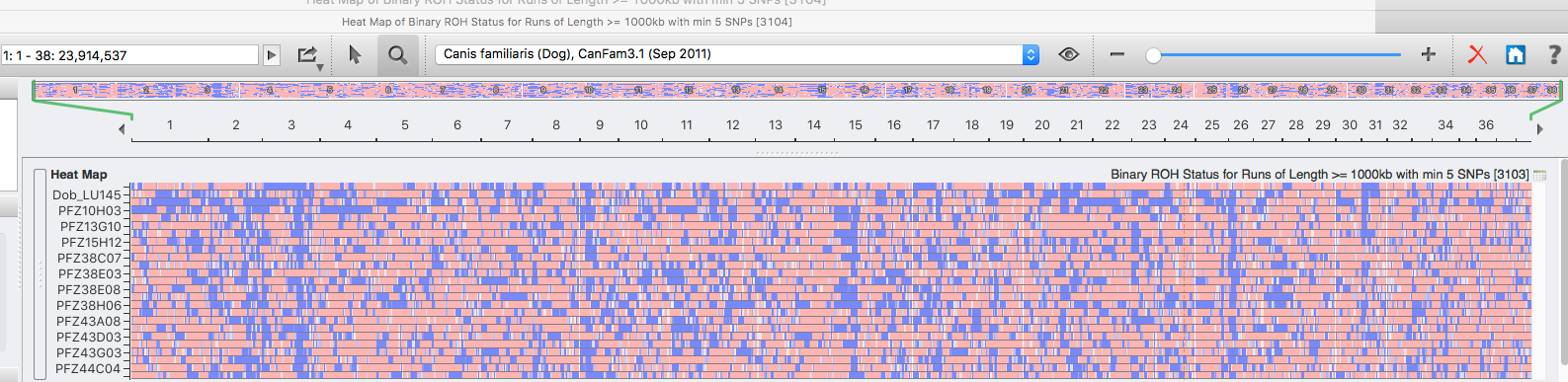
| For the 6 figures below: Inbreeding computed from pedigree data for 5, 10, and all generations for (top to bottom): Papillon, Bernese Mountain Dog, Golden Retriever, Basenji, Norwich Terrier, and Nova Scotia Duck Tolling Retriever. With this ordering, the breed with the least current level of inbreeding is first, and the others follow in order of increasing magnitude (i.e., the highest current inbreeding computed from pedigree data is for the NS Duck Tolling Retriever). |
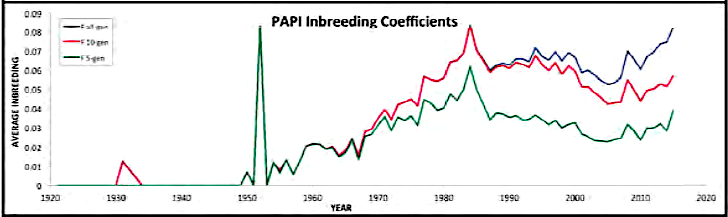
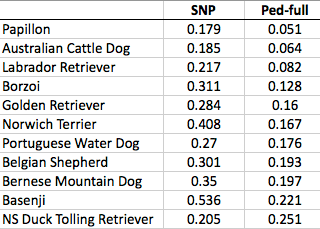
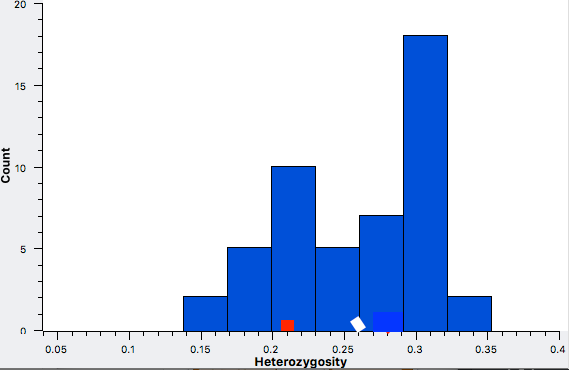
These data confirm that inbreeding of most purebred breeds is extremely high, with all but a handful of values exceeding the level of inbreeding produced by mating full siblings from unrelated parents.
ICB's online courses
*******************
Join our Facebook Group
ICB Breeding for the Future
...the science of dog breeding
*******************
Visit our Facebook Page
ICB Institute of Canine Biology
...the latest canine news and research