How much genetic diversity is there in your breed? How can you find the dogs that might have the genetic variation that you're looking for?
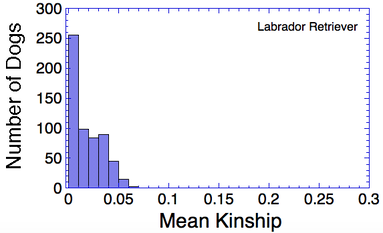
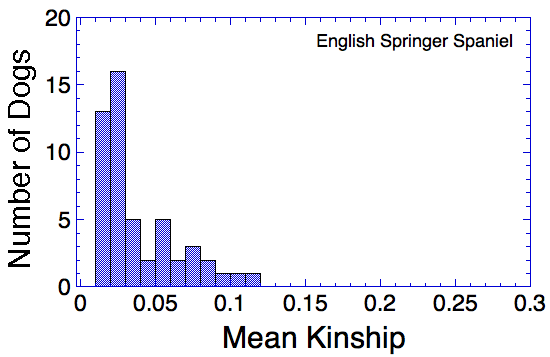
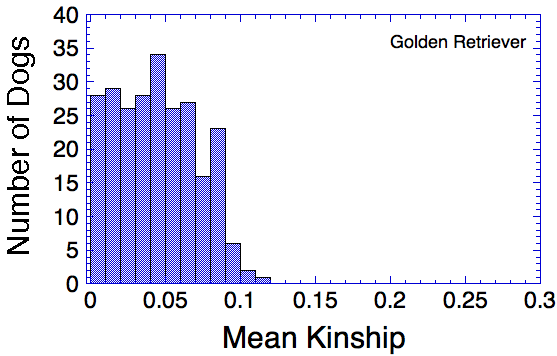
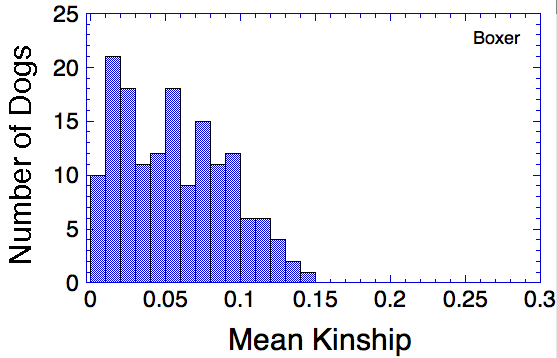
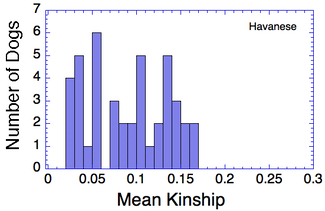
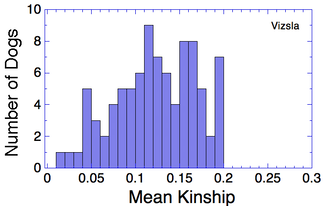
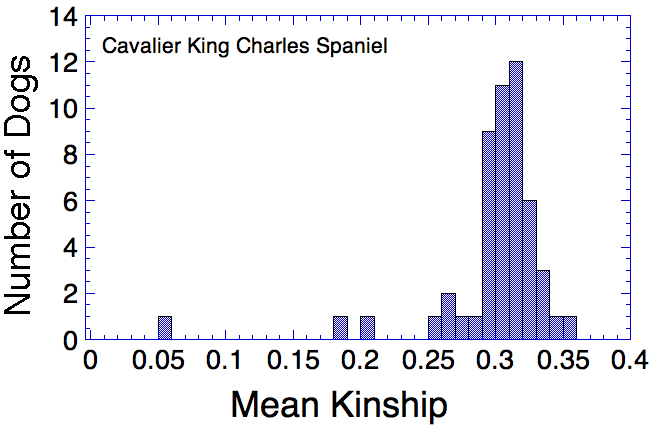
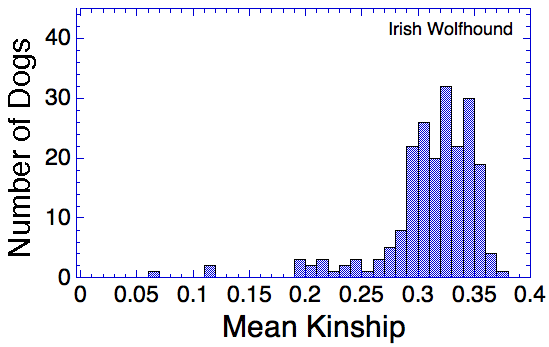
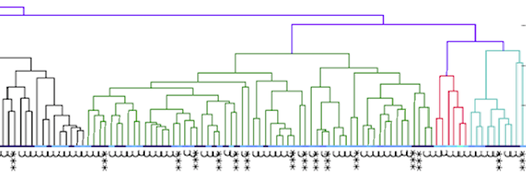
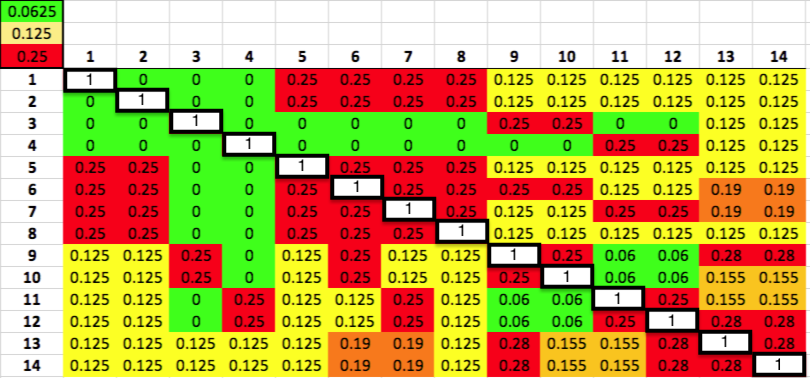
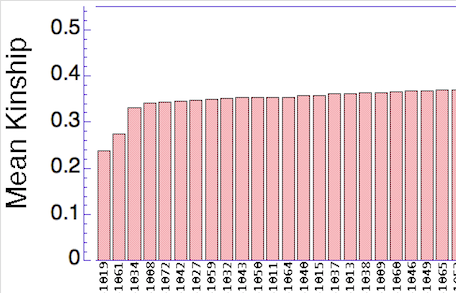
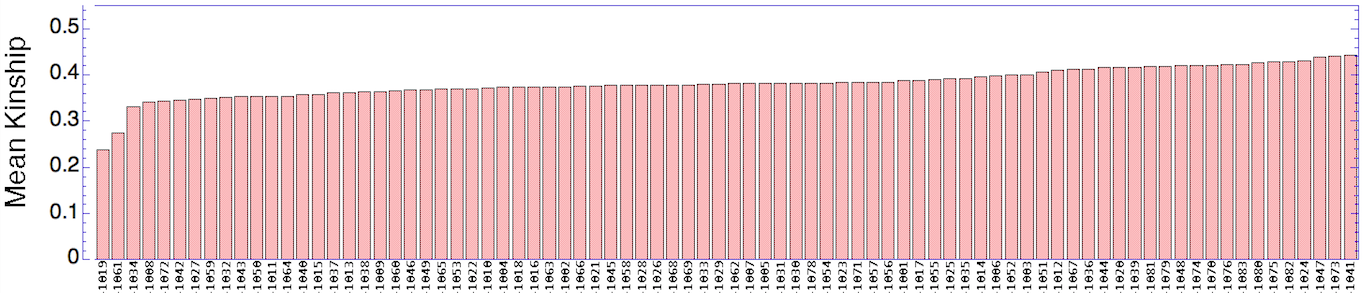
Let's say that we have a population of animals that are not closely related. If we compare the genes in each dog with those in every other dog in the population, we can compute each dog's mean kinship, which I explained in a previous post. Since we know the dogs in our population are relatively unrelated, we know that the values for mean kinship will be low (they can range from 0 to 1). If we graph the mean kinship of all of the dogs in the population, we might get a chart that looks like the first one on the left below for Labrador Retrievers, in which all of the values are less than about 0.06. On the other hand, if we have a population of closely related dogs, we might get a graph like the last one on the right below for Irish Wolfhounds, in which most of the values are greater than 0.3. If you scan the graphs for the other breeds depicted below, you will see that the distribution of values for mean kinship can vary considerably.
Remember that mean kinship is the average relatedness of a dog to all others in the population. The Labrador Retriever is one of the most popular breeds in the world so numbers are very high, and there are populations of show dogs, hunting dogs, personal pets that will all be a bit different from each other genetically, and populations in different countries will probably be different from each other as well. This is a breed with a LOT of genetic diversity, and the mean kinship values are mostly low. But if you were to make another graph with just the show dogs, the distribution would probably be skewed towards higher values because those dogs would be more closely related to each other than they are to field line dogs.
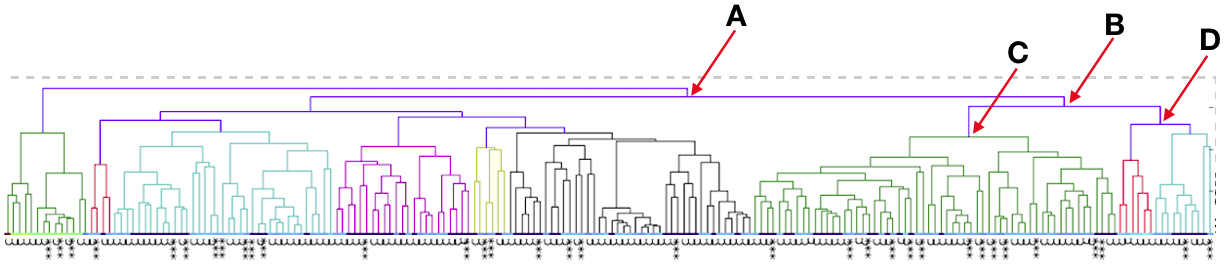
When you compare Labradors with Irish Wolfhounds, the graph is skewed strongly towards much higher values. This indicates in this population of over 200 dogs, the average relatedness was very high. The exception is a few dogs that fall far below the pack and are not closely related to the bulk of the population. This indicates that most of the dogs are closely related to each other.
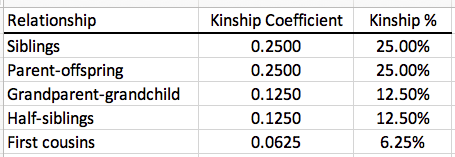
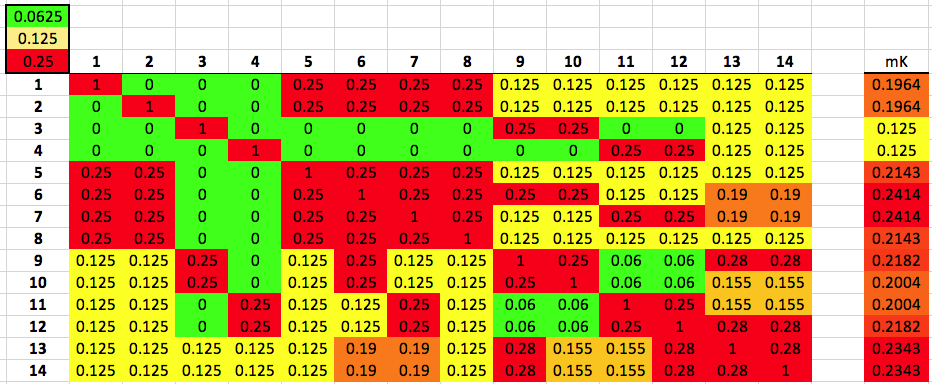
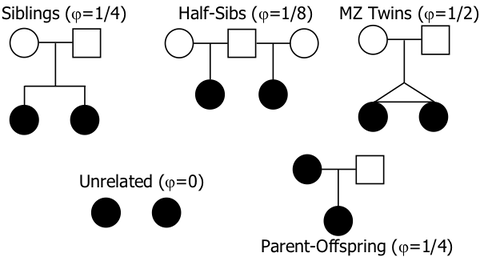
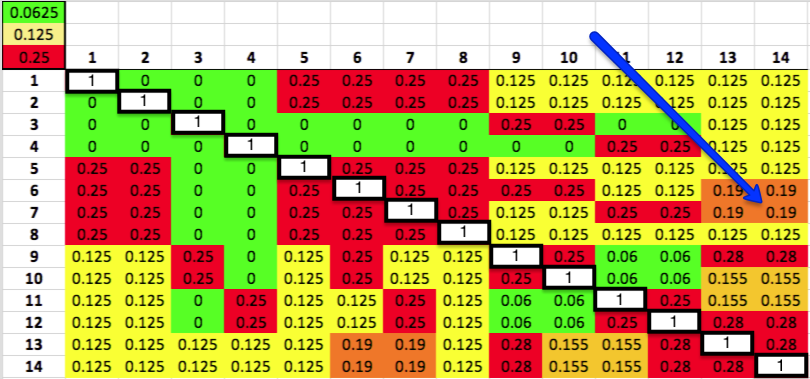
You can do the same thing for the graphs for other breeds, using this chart for the kinship coefficients for each level of relatedness.
So, looking at the chart for Irish Wolfhounds (bottom right), you can see that most of the dogs have a mean kinship > 0.25; that is, on average most of these dogs are more closely related to each other than full siblings. Imagine a family reunion with a few aunts, uncles, and cousins, and 134 of your siblings. This is a breed with a very high level of average relatedness to each other. Likewise for Cavalier King Charles Spaniels. In the small sample of Havanese, there two clusters of dogs, one with a relatively high level of relatedness to each other, and another with rather low relatedness.
pedigree and DNA data using the ICB Breeder Tool.
Are you interested in using the ICB Breeder Tool with your breed?
Contact us to get started!
Cool tricks with Kinship Coefficients, part 1: "Is this dog really an outcross"?
Cool tricks with Kinship Coefficients, part 2: "Should I breed this dog?"
Cool tricks with Kinship Coefficients, part 3: "How can I manage a disease without a DNA test?"
ICB's online courses
***************************************
Visit our Facebook Groups
ICB Institute of Canine Biology
...the latest canine news and research
ICB Breeding for the Future
...the science of animal breeding