So what do you do? How can you identify a dog that will be a good outcross for your breeding program?
|
Conservation genetics has the perfect tool for this. It's called the "kinship coefficient" (K), and it measures the genetic similarity between two individuals (Li et al 2011).
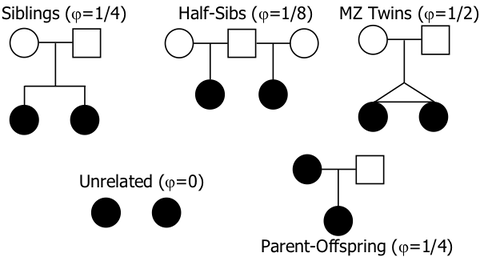
In particular, the kinship coefficient tells you how much of the genomes of two dogs are the shared because they were inherited from a common ancestor. (You will also see the kinship coefficient referred to as the "coancestry coefficient"; they are the same.) The kinship coefficient reflects shared ancestry (coancestry) in the same way as the coefficient of inbreeding. First cousins will, on average, share 6.25% of their genomes, and K for the pair will be 0.0625. In half-siblings, the prediction of fraction shared is 12.5%, or K = 0.125; and for full siblings K = 0.25, or 25%. |
The kinship coefficient can be calculated from a pedigree database, just as inbreeding coefficients are. But it can also be estimated directly from DNA data - in effect, the "realized" kinship coefficient that reflects the actual level of shared ancestry between two dogs based on their DNA.
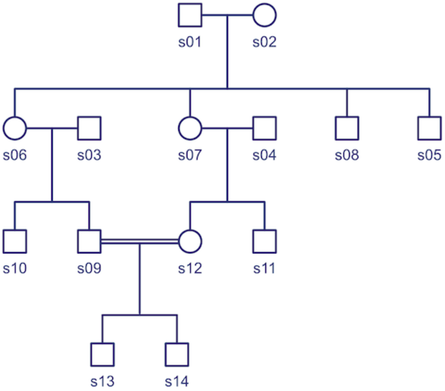
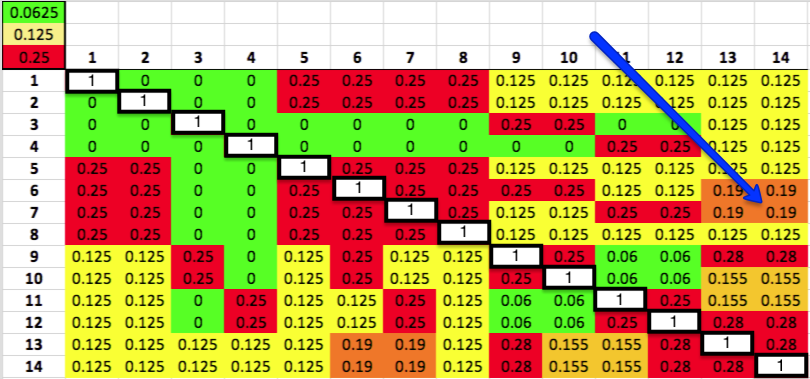
The kinship coefficient is always a comparison of two dogs. You can compute the K for each pair of the dogs in the pedigree and display them in the form of a matrix. Each dog is listed down the side and also across the top. The number in the box where the column and row of a pair of dogs intersects is the kinship coefficient of that pair. The white boxes on the diagonal are each dog compared with itself.
For example, we can determine that the kinship coefficient between dog 14 and dog 7 is 0.19, or 19%. This means that if you were to choose one allele at random from the pair of alleles at a particular locus on the DNA of dog 14, and likewise choose an allele at random from that locus for dog 7, the probability of choosing two alleles that are identical because they were inherited from a common ancestor is 0.19, or 19%. To put it another way, the chance that the offspring of this pair of dogs will be homozygous at that allele (have two identical copies of an allele inherited from a common ancestor) is 19%. Furthermore, if these two dogs were to produce a litter, the predicted coefficient of the puppies would average 19%.
Computed from a pedigree, the kinship coefficient is an estimate of the average expected kinship coefficient; that is, the shared genetic relatedness because of shared ancestry. But kinship can also be determined directly from DNA genotyping data, such as is provided by Embark. (As far as I know, this is the only company that will allow dog owners to download the raw data for their dogs.) When based on DNA, these are "realized" rather than predicted estimates of genetic relatedness, and they will be different for every dog in a litter.
To answer these questions, you can do direct comparisons of the genomes of these two dogs with the bitch you want to breed by calculating the kinship coefficient for both of the sire-dam pairs. Modern DNA analysis using "single nucleotide polymorphisms" (SNPs) can now provide excellent data with very high resolution that you can use to do this. ("Single nucleotide polymorphism" is a fancy term for loci that are highly variable ("polymorphic"); the abbreviation is pronounced "snips".) We can easily and inexpensively compare hundreds of thousands - even millions - of loci on the chromosomes of a pair of dogs, assess whether the alleles at each of those loci are the same or different. Further, using data for allele frequencies in the larger population we can determine if the shared SNPs are copies of the same allele in a common ancestor; that is, they are "identical by descent" (IBD). This is the technique used for establishing the relationships among many breeds of dogs (Dreger et al 2016), and it is also used to identify genetic mutations associated with diseases in genome-wide-association studies (GWAS).
Instead of estimating kinship from a pedigree database, which will only give you one number that applies to the entire litter, you can estimate the actual degree of genetic similarity between two dogs. In fact, using SNPs you can even see which areas on each chromosome are shared. You can use this information to see if blocks of homozygosity overlap in two dogs, in which case all of the offspring would also be homozygous in the same place. (More on this in an upcoming blog post!) You can specifically look at characteristics of the genome of the two litter mates and determine if there are any ways one might be a better match to your bitch than another. And the kinship coefficient will also tell you the estimated inbreeding coefficient of the litter produced by each potential sire.
For the preservation breeder, the genomic kinship coefficient from SNP genotype data provides you with the best, most advanced tool available today for genetic management to improve the quality of your puppies while also protecting genetic diversity for the health of your breed in the future.
We will be learning about kinship coefficients and how to use the kinship matrix in ICB's new course, "Strategies for Preservation Breeding", which starts 10 September 2018. Join other preservation breeders who will be using this powerful tool to improve their breeding program. If you're interested in using genomic kinship coefficients to explore options for your next breeding, contact ICB and we will work with you to set that up.
Watch this space for more Cool Things you can do with kinship coefficients!
Dreger, DL, M. Rimbault, BW Davis, A Bhatnagar, HG Parker, & EA Ostrander. 2016. Whole-genome sequence, SNP chips and pedigree structure: building demographic profiles in domestic dog breeds to optimize genetic trait mapping. Disease Models and Mechanisms 9: 1445-1460. (pdf)
Hayward, JL, MG Castelhano, KC Olivera, and others. 2016. Complex disease and phenotype mapping in the domestic dog. Nature Communications 7:10460. DOI: 10.1038/ncomms10460. (pdf)
Li, M-H, I Stranden, T Tiirikka, M-J SevonAimonen, & J Kantanen. 2011. A comparison of approaches to estimate the inbreeding coefficient and pairwise relatedness using genomic and pedigree data in a sheep population. PLoS ONE: Nov 2011; Vol 6:11. e26256. (pdf)
this terrific new ICB course!
|
|
ICB's online courses
***************************************
Visit our Facebook Groups
ICB Institute of Canine Biology
...the latest canine news and research
ICB Breeding for the Future
...the science of animal breeding