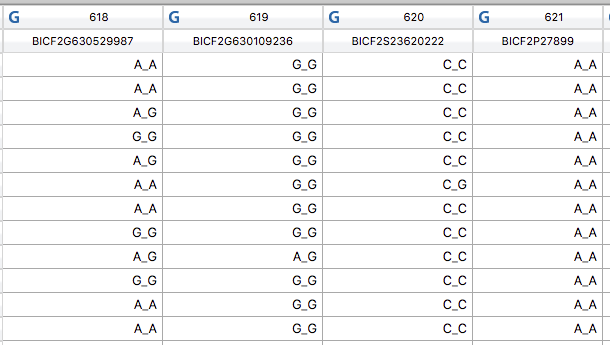
Our ability to resolve genetic relationships among individuals has been revolutionized by technology that can identify hundreds of thousands, or even millions, of DNA markers. Distributed densely over all of the chromosomes, these markers, called SNPs (pronounced "snips"), can reveal present genetic relationships in fine detail. Even more amazing, SNPs can tell us about genetic history - what carnivores are the dog's closest relatives, where the world's dog populations originated, what breeds were used in the creation of new breeds by crossbreeding, and even how long in the past a cross-breeding occurred. The amount and quality of information is breathtaking, and the study of canine genomics has been changed forever. This SNP technology is referred to as "Next Generation Sequencing" (NGS), because it truly did propel genomics into the "next generation".
We can use the power of high resolution SNP analysis to reveal the details of genetic relationships among animals. A potential sire that might look like an outcross to your bitch on paper can instead be very similar genetically because of relationships among ancestors many generations back in the pedigree. When you are choosing a sire to improve a trait, reduce the risk of genetic disease, or increase the genetic diversity in your line, reliable information about the DNA in the dogs can be the difference between success and failure. Let's look at an example of how we can use information about genetic relationships to manage a genetic disorder in dogs.
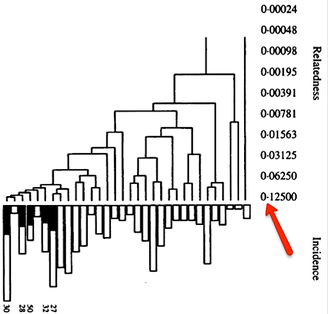
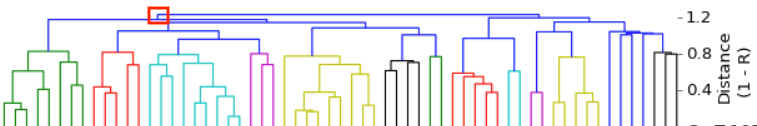
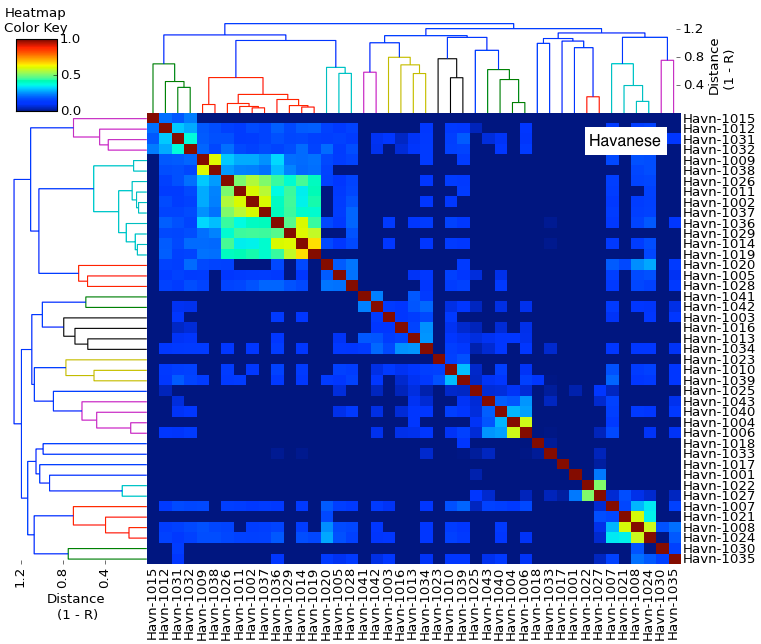
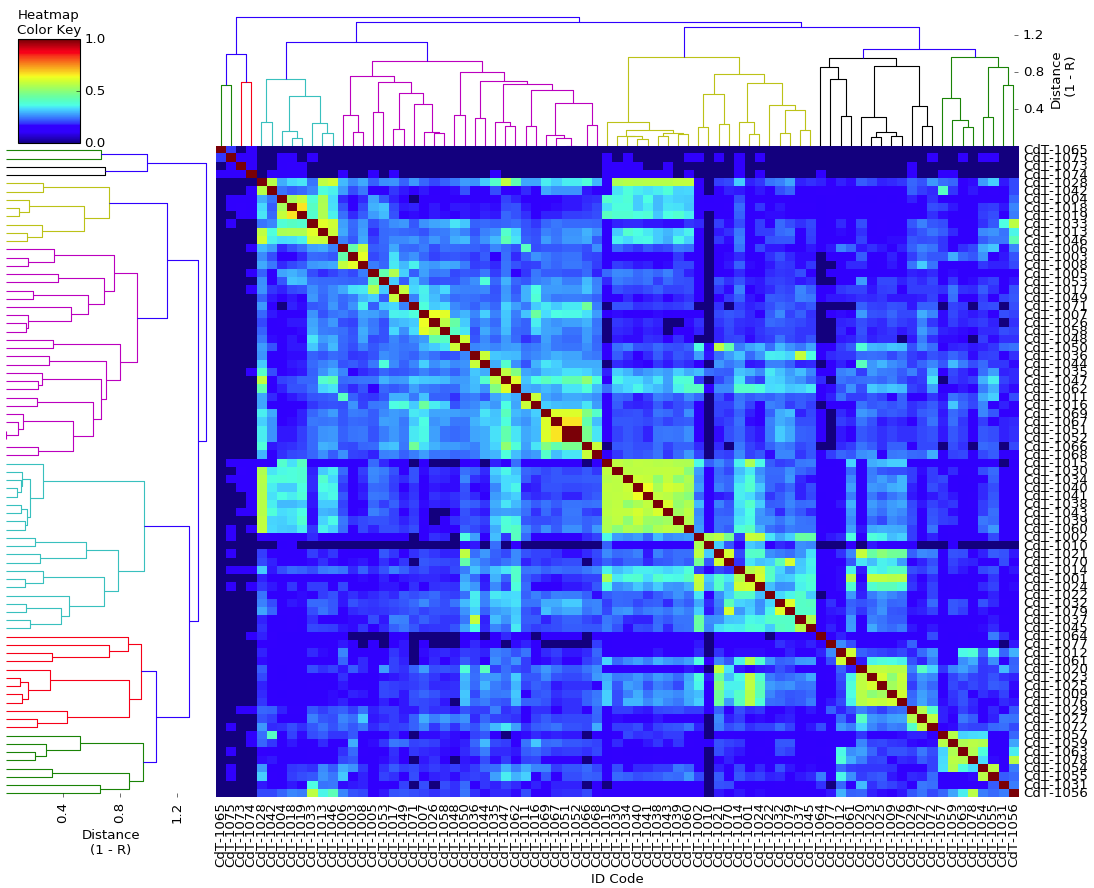
| After creating this map of the relatedness of the dogs in their breeding colony, the number of dogs with ED was indicated by shading the bar of that group. This clearly revealed that the dogs with ED were closely related and clustered together in one region of the map. Using this information based on genetic relatedness, ED could be reduced by identifying the unaffected dogs that were at high risk because they belonged to genetic clusters containing affected dogs - and without knowing anything at all about the genes involved or mode of inheritance. This analysis was done in 1998, not using a fancy modern DNA test, but a carefully-tended pedigree database. The limitation of using pedigrees for the general population of purebred dogs is that they are often incomplete, they can (and surely do) contain errors, and often are not even available. |
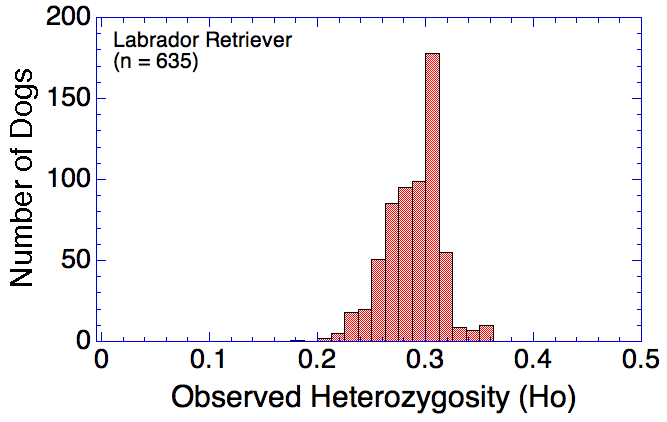
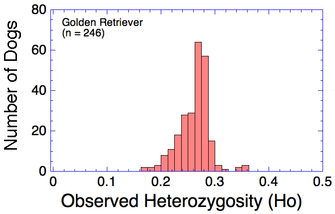
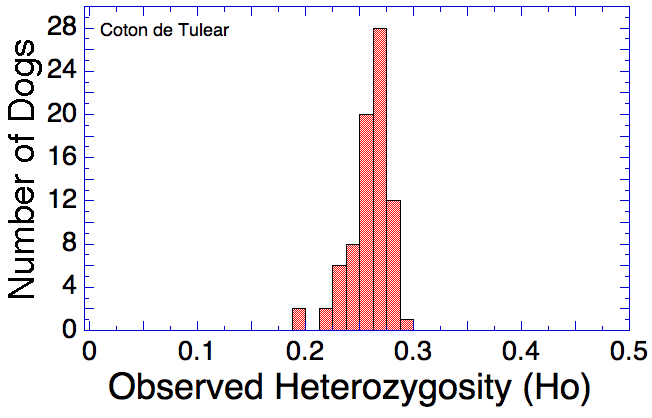
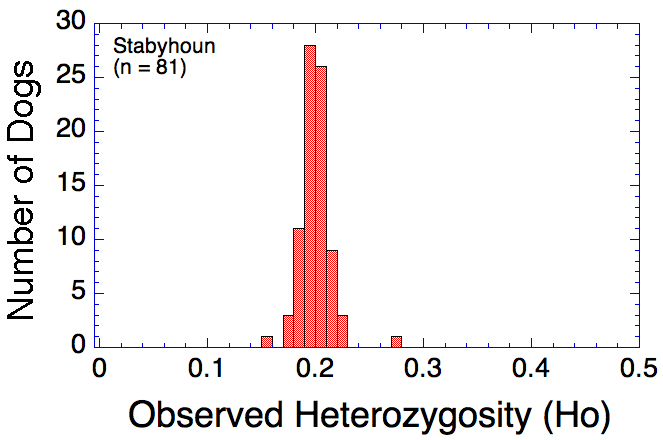
Many breeds are highly inbred and have low genetic diversity. Will this technique using DNA to establish genetic relatedness work for purebred dogs?
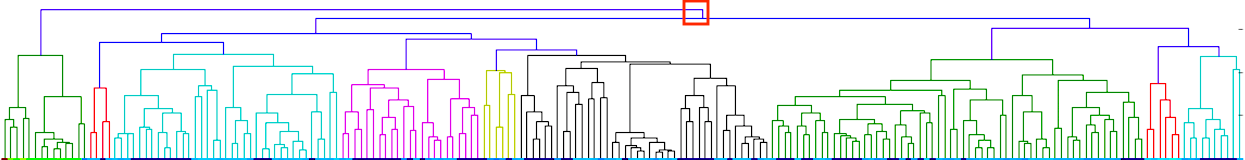
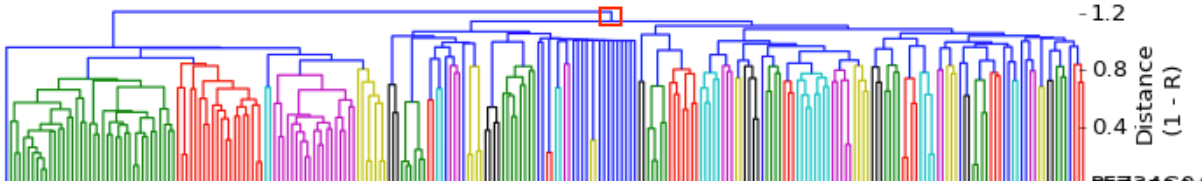
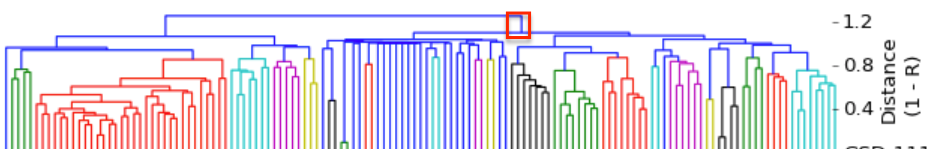
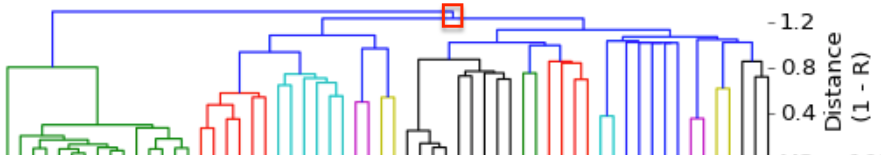
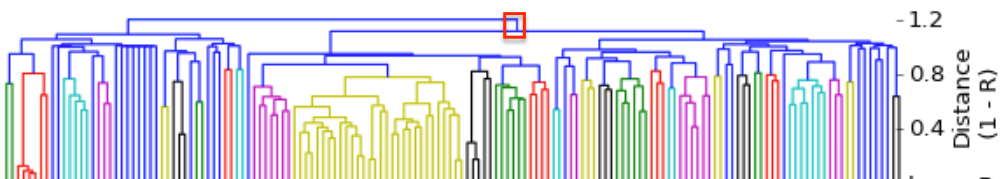
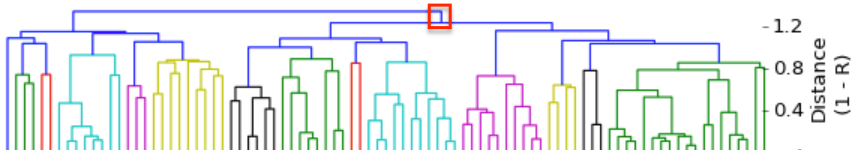
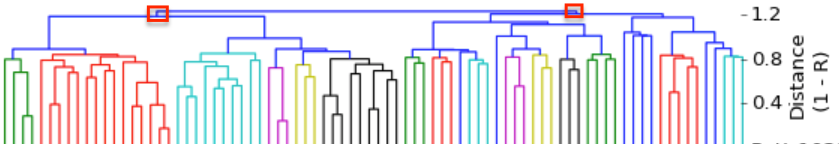
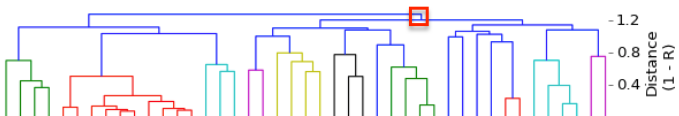
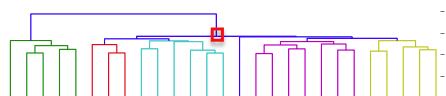
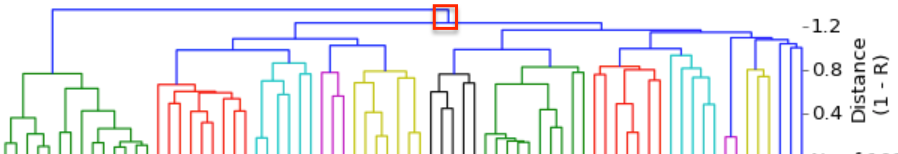
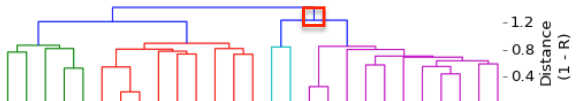
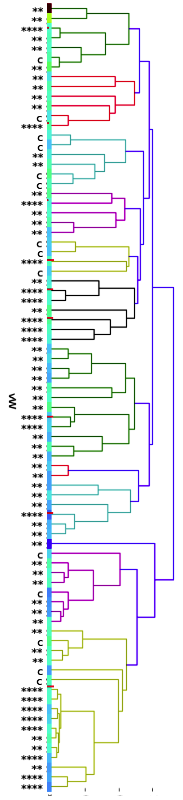
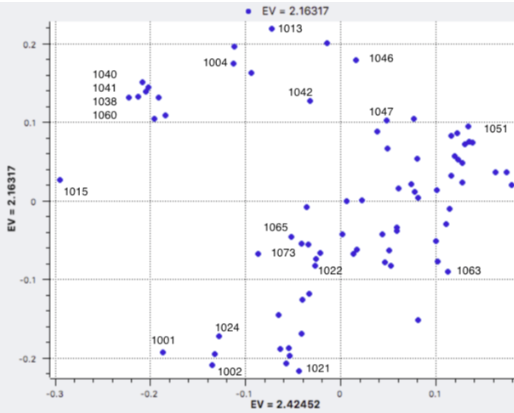
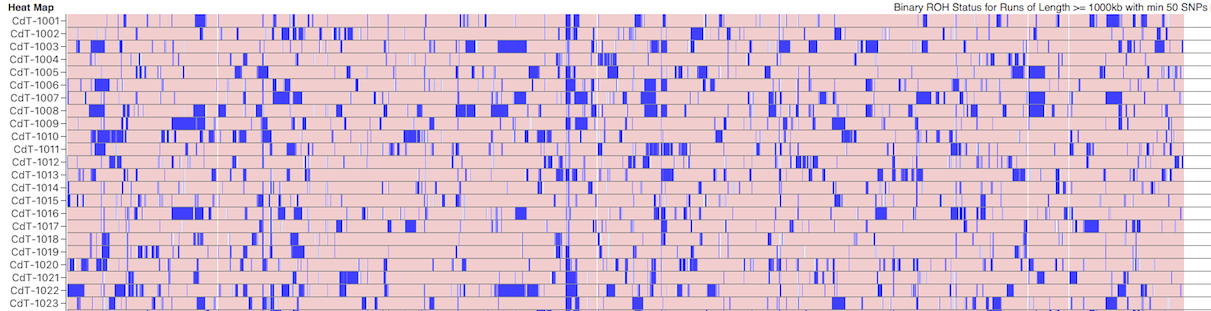
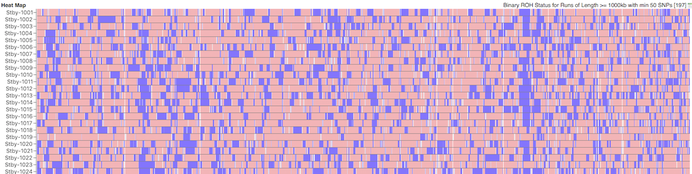
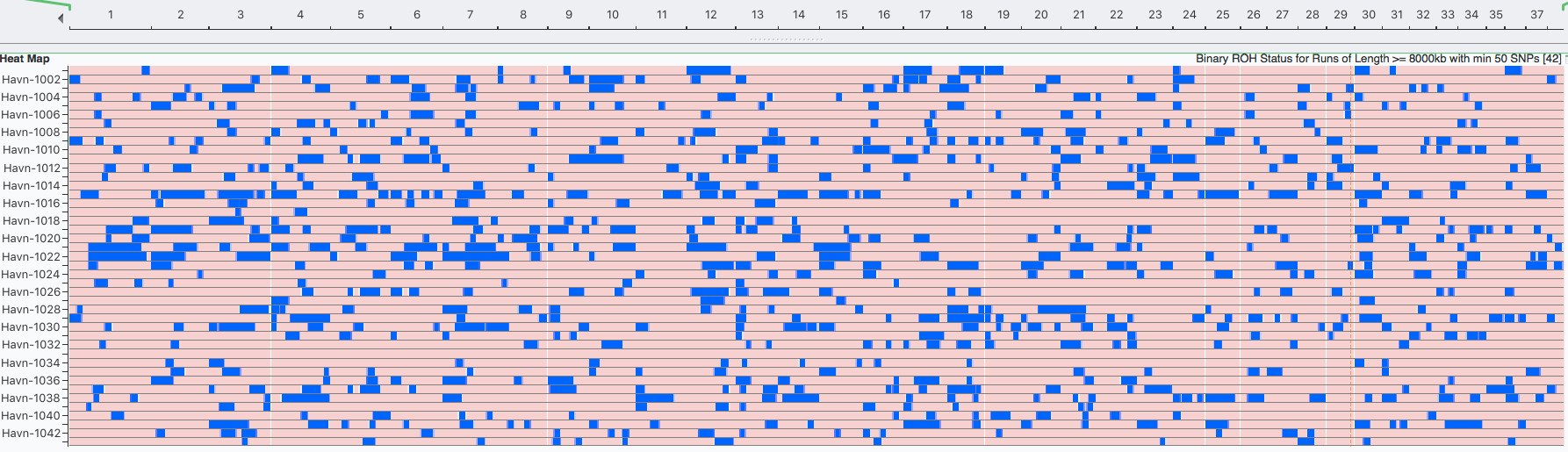
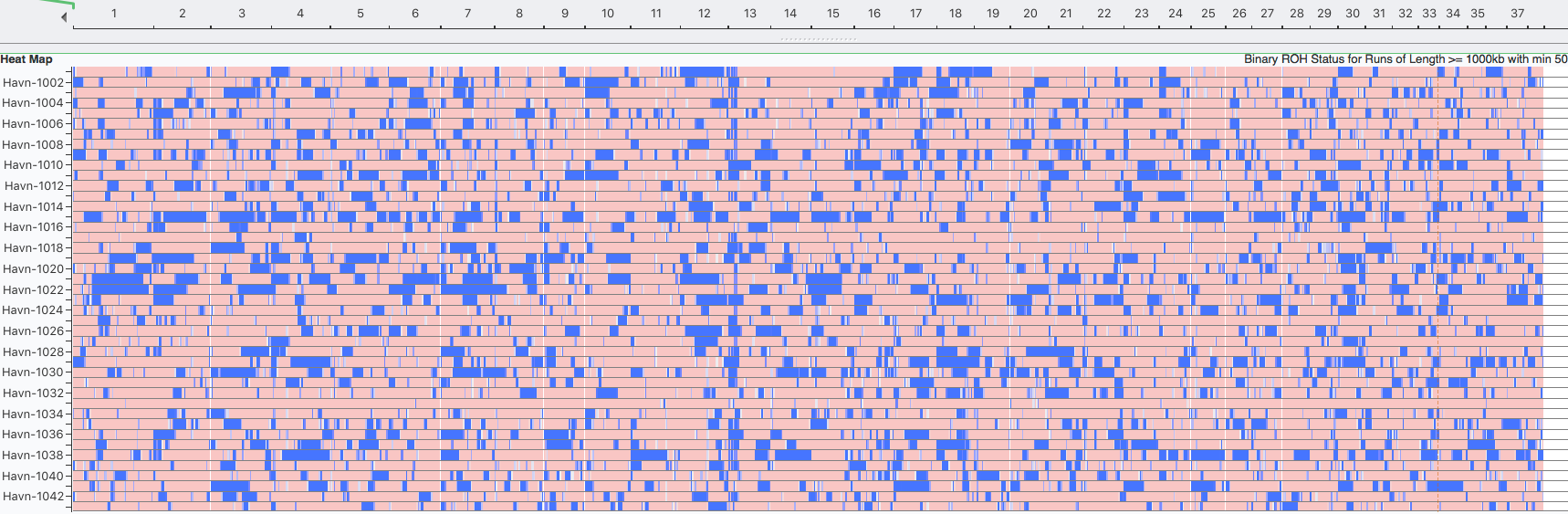
I have gathered some DNA data in public archives from published studies that were analyzed using high density SNP genotyping, then used cluster analysis (as was done above) to produce the map of genetic relatedness for a number of breeds. These are displayed below, and many show that there are genetically distinct subpopulations within a breed that could be exploited to look for patterns in the expression of genetic disorders. We know nothing about who these dogs are, but the charts show that there is sufficient genetic differentiation in a breed to use for this type of analysis. On each one I have indicated with a red square the first major division to make it easier for you to visualize the groups. For some of these we clearly need more data, and for all of them we need data for known dogs. But this should give you a good idea of what some of these look like.
Here we describe a very powerful technique to sort out genetic relationships among individuals that can be used to improve genetic management of disorders without knowing the genes involved. There is a long list of inherited disorders that breeders are struggling to manage in the absence of genetic information for guidance. Epilepsy, cancer, heart disorders, kidney disease, neurological problems, degenerative myelopathy, and the list goes on and on. We're not making much progress.
We can do this now. We need DNA genotype data for current dogs, including those in the breeding population, to establish the genetic subgroupings within the breed. We need the incidence data for the disorders of interest.
It might not take a lot of data to reveal the information we need. In the example of Laboradors above, data from just a few dogs would indicate the location of the cluster containing the affected dogs. You could take action using this information far sooner than if you needed 20 affected and 20 clear dogs to search for genes.
ICB's online courses
***************************************
Visit our Facebook Groups
ICB Institute of Canine Biology
...the latest canine news and research
ICB Breeding for the Future
...the science of animal breeding