But Mom is no fool. "Billy", she says sternly, "eat your peas instead of just pushing them around on the plate!" Busted.
We risk doing a similar thing when making decisions about the ideal sire and dam for the next litter of puppies. Let's say you have a bitch you want to breed and you're considering four or five potential sires. You also have some DNA test information that allows you to compare the genetic similarity of the bitch with each of the sire candidates. Selecting the sire that is least similar to the dam should produce puppies with the lowest average inbreeding.
But does this tell you everything you need to know? Are you really improving genetic diversity? Or are you just pushing the peas around on the plate?
Let's consider 2 things. First, let's look at genetic relatedness.
By "relatedness", we refer to the genetic similarity of a dog to the other dogs in the population. Closely related dogs will be more similar; distant relatives will be less similar.
There will always be genetic differences between two dogs, no matter how closely related they are, because no two puppies will inherit exactly the same alleles out of all the zillions of possible combinations. So even among closely related dogs, you should always be able to identify a dog that is "least" related, and if you're shopping for sires this one might look like the best bet.
The problem here is that you can identify a dog that is less related compared to the others, but it might still be very closely related - much more closely related than you would risk breeding to if you knew the actual genetic level of relatedness (First cousins? Half-siblings? Full siblings???).
Let's consider an example. We have a population of about 80 dogs in a particular breed, and you are sorting through them to identify which of the sires you are considering is least similar to your bitch. You make your choice and since they don't share any common ancestors for several generations, you're happy.
But the kinship coefficients of the dogs would tell a different story. The kinship coefficient for a pair of dogs is the fraction of their genes that are identical by descent. This sounds like it's related to inbreeding, and it is. In fact, the inbreeding coefficient of a dog (the fraction of the genes in an individual that are identical by descent) is equal to the kinship coefficient of its parents. Since we know that most genetic disorders in dogs are caused by recessive mutations, the kinship coefficient also tells us the risk of producing a genetic disorder caused by a recessive mutation in the puppies.
If you determine the kinship coefficients between the bitch and each of the potential sires, you choose the one with the lowest kinship coefficient. However, the kinship coefficient reveals that this "least related" dog is actually more similar to the bitch genetically than you would expect of a full sibling. The data for the kinship coefficients with these potential sires are:
| 1) 0.388 2) 0.693 3) 0.340 4) 0.398 5) 0.288 |
Wow. You wouldn't do a full sibling mating, and this mating would be a similarly close inbreeding. From the data for the other sires, it looks like there is very high relatedness among many of the dogs in the population, whose kinship coefficients go as high as 0.693!
When you compared the dogs relative to each other, one dog looked like a good choice. But after looking at the actual kinship coefficients of the potential parents, you decide that the marriage is much too close for comfort.
The second issue comes from the first. Genetically similar dogs will have similar mutations. The least related pair based on kinship coefficients will nevertheless share many mutations if they are as similar as full siblings. They might be from different lines and not share any kin for many generations, but genetic similarity brings with it high risk of producing a genetic disorder. Mating of full siblings, with a kinship coefficient of 0.25, has a 25% risk of pairing up two recessive mutations.
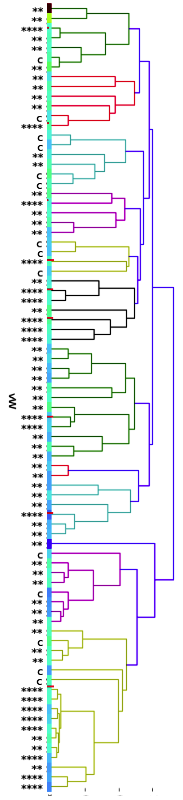
Let's look at another example using this same breed. Below is a "dendrogram", a family tree that is based on kinship coefficients determined using a high-density (> 200,000 markers) SNP panel that has considerable power for resolving relatedness among closely related individuals.
On the panel on the left, I have indicated the results of genetic testing for the mutation (VWF) responsible for von Willebrand's disease in this particular breed. Individuals that tested clear have a little "c" at the left end of the branch. Individuals that are carriers of the mutation have two asterisks (**), and dogs that are homozygous (and therefore affected by the disorder) are indicated with four asterisks (****).
You should be able to scan from the top of the chart to the bottom and easily pick out the clear, carrier, and affected dogs. What you will probably notice is that the VWF mutation is widespread in this breed. If there wasn't a test for this disorder, you might think you're avoiding it by choosing dogs that are not closely related to each other. But, as we saw above, all of these dogs are very similar genetically, regardless of how unrelated they might appear on paper.
Considering the more complete information for genetic similarity provided by the kinship coefficients, together with the DNA test results, you can see that you aren't really going to improve the genetic diversity in your litter, no matter what you do. You can make all the comparisons and choose what looks like the best one option, but it's really just pushing the peas around on the plate.
To learn more about the genetics of dogs, check out
ICB's online courses
***************************************
Visit our Facebook Groups
ICB Institute of Canine Biology
...the latest canine news and research
ICB Breeding for the Future
...the science of animal breeding