In dogs, we often breed only one or two puppies from a litter, selecting those that we feel best suit the goals of the breeder and are good representatives of the breed. But if you started with a sire and dam that you felt were quality dogs, all of those puppies should have "quality" genes but in different combinations, some of which make for a "better" puppy than others. When the pups judged to be less suitable are not bred, we risk losing from the gene pool some of the genes that produced the good quality in the parents. We limit our future breeding options when we lose those genes, and we also remove the opportunity of tossing them together with a mix of genes from another parent dog to perhaps produce something wonderful. You can't breed every dog, and not every dog is worth breeding.
If you breed for both quality (on whatever scale) and health (and of course you do, right?), you should be concerned about the inadvertent loss of genetic diversity that results from selective breeding because eroding the genetic base of the breed will have consequences down the line. But how do you determine which dogs are important for preserving genetic diversity? You can use the kinship coefficient.
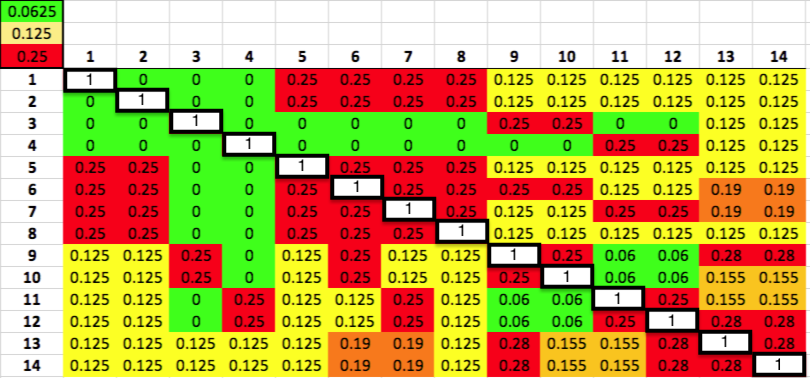
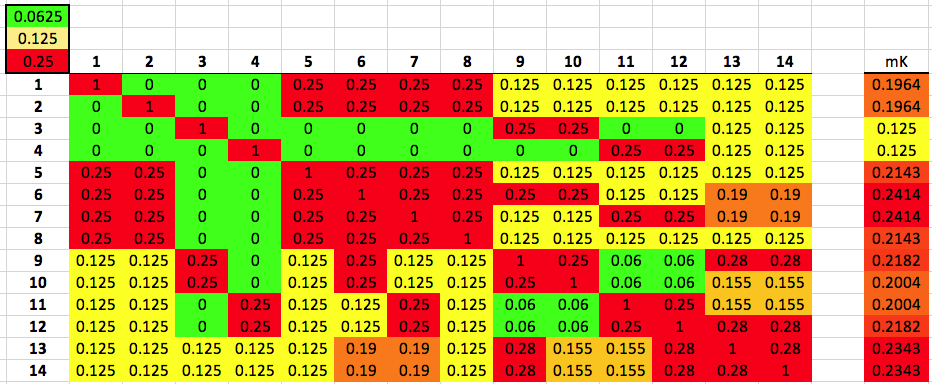
Below is the same kinship matrix we talked about in my previous post about kinship coefficients. Again, the white squares on the diagonal are each dog compared to itself (so K = 1), and the other squares are color-
coded to make it easy to spot combinations that are less (or more) related to each other.
We can use the color coding indicated in the three squares in the upper left corner on the matrix to easily spot dogs that are closely related to other dogs (i.e., green, yellow, and red). If you follow across the rows of dogs 6 and 7 from left to right (or follow down the columns for these two dogs if you prefer), you will see that there are lots of red, orange, and yellow squares, indicating dogs as closely related to 6 and 7 as half-siblings (K = 0.125) to full siblings (K = 0.25). This means that the genes in dogs 6 and 7 are also found in other dogs in the population because they share ancestry.
Now do the same thing for dogs 3 and 4. You will find that most of the squares are green, indicating that they are not closely related to most of the dogs in this group. Interestingly, 3 is as related as a full sibling to 9 and 10 (K = 0.25), which are likewise closely related to each other; similarly, 4 is closely related to 11 and 12, which are likewise as related as full siblings. It would be interesting to look at the genealogical relationships among these four dogs from pedigrees.
If dogs 6 and 7 have many relatives in the population and dogs 3 and 4 have very few, then the latter pair are genetically more valuable because they carry genes that are not common among these dogs. It is fairly easy to identify the genetically important dogs in this small example population, but how can we do it with a large population of dogs with complicated patterns of relationship?
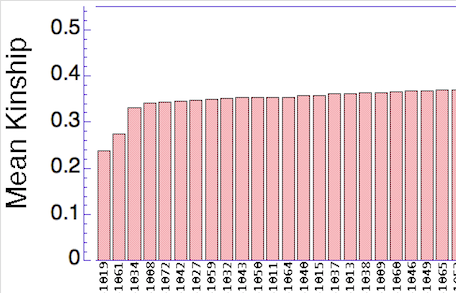
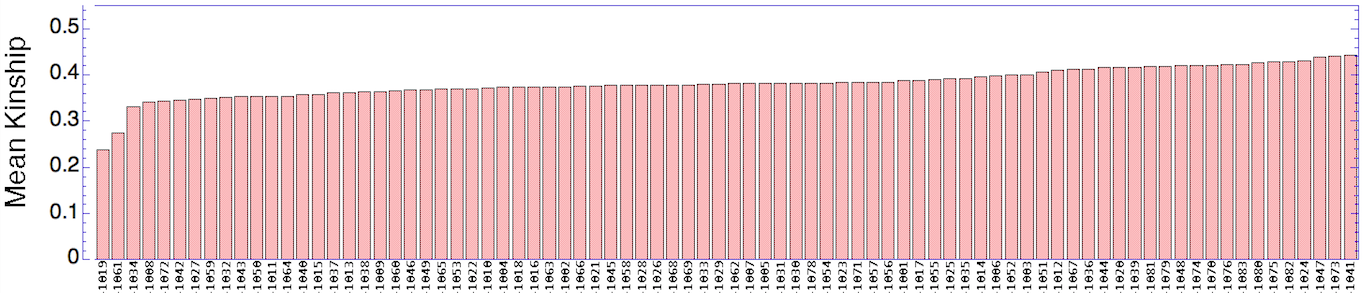
Now we can use the data for mean kinship to quickly identify the dogs that are genetically most valuable in a large population. The simple bar graph organizes the dogs by rank, with the lowest values of K to the left - the dogs with the highest genetic value - and high values to the right (the dogs with lowest genetic value). In this population, there are a few dogs with high genetic value, but most of the rest of the population is closely related.
We should be using kinship coefficients much more than we are, and that is to the detriment of maintaining genetic diversity in our breeds. Breeders should know which dogs are genetically valuable, even if they aren't the top winners in competition, because the genetic variation they carry is the raw material for breed improvement in the future. Lines lost are lost forever. Perhaps breed clubs should have a genetic diversity committee that is responsible for monitoring the population to prevent loss of variation, or this could fall to the health committee because diversity and health go hand in hand. Note that if everybody rushes to breed to the least related dogs, their genes will become common and intermixed in a way that prevents them from being used strategically to manage genetic diversity. Diversity should be exploited strategically. (For the same reason, there should be a "popular sire" monitor, whose job it is to keep an eye out for sires producing more than their fair share of puppies.)
Learn how to use the kinship coefficient to take some of the guesswork out of decisions about genetic relatedness or uniqueness when you are considering your future breeding plans. This simple tool can provide you with a wealth of valuable information!
this terrific new ICB course!
|
|
ICB's online courses
***************************************
Visit our Facebook Groups
ICB Institute of Canine Biology
...the latest canine news and research
ICB Breeding for the Future
...the science of animal breeding