It seems you can never know enough about genetics. On a regular basis science tells us about a new wrinkle that really messes up what we think we know. If you're a biologist, this can be really exciting, but if you're a dog breeder it can create a lot of discomfort. We're just now realizing that as autonomous as you might think you are, your bacteria are really in charge, making you smarter, sicker, healthier, or smellier, and affecting your hormones, behavior, and immune system. (There is a nice review HERE.) And very recently, we've discovered that they affect the inheritance of traits from mother to offspring in really creepy ways.
While we're worrying about keeping up with the pace of new discoveries in genetics, there are some genetic fundamentals that don't get their fair share of attention. One of these is something called "genetic drift". Jeffrey Bragg, who has been breeding Seppala Siberian sled dogs for many years and knows a thing or two about genetics, calls it "the breeder's hidden enemy" - pretty strong words for something most breeders never think about. (You can read some of excellent essays by Jeffrey on his website, or you can download a collection HERE.)
So what is this insidious force that we should be worrying about? It's a simple concept, and it is described well by the name. Genetic drift is the fluctuation in the frequencies of alleles in a population caused by random chance. There are two alleles at each locus, and inheriting one or the other is like flipping a coin; if you flip a coin 1000 times, you should get pretty close to half heads and half tails. But if you flip the coin only 5 times, you could get - just by random chance - all tails, and if you try it again you might get all heads. If the coin is "fair", the frequency of heads (or tails) should get closer and closer to 50% the more tosses you do.
This phenomenon - the random variation in frequency cause by chance selection of one outcome or another - is the basis of genetic drift. As chance might have it, if one of a pair of alleles gets passed to more offspring more frequently than the other, that allele will have a higher frequency in the next generation. In the next generation, the other allele might do better, and the frequency of that one will rise and the other one will fall. It can happen that, just by chance, the very last copy of an allele can be lost due to genetic drift and the entire population is then homozygous for the other allele. Of course, breeders have no control over which allele of a pair is inherited by a puppy, so genetic drift pushes allele frequencies around in a population just out of sight of the breeder, who is likely completely unaware.
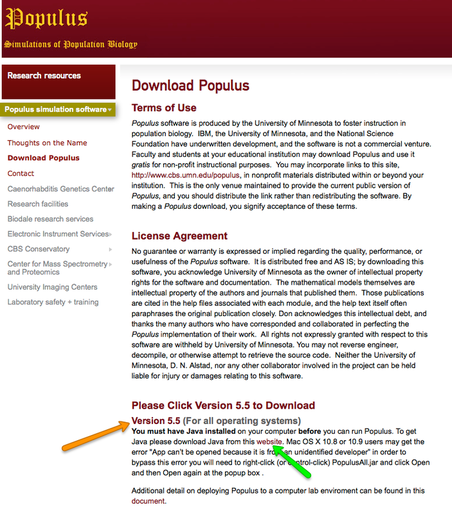
To give you an idea of how genetic drift works, there is a clever little computer simulation that will let you do some experiments of your own. The software is called Populus and is a Java routine that will run on both a Mac and PC. Here's how you get it:
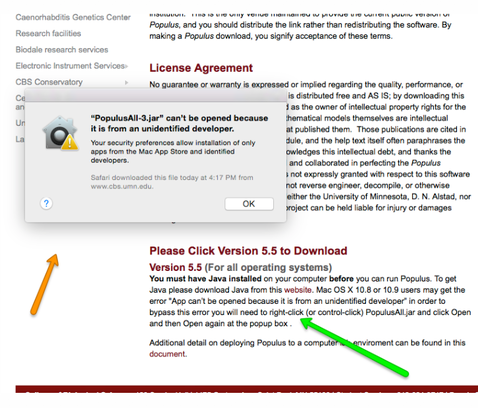
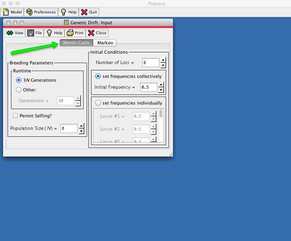
Double click on the downloaded file and it should open to the program, but you might instead get this warning box complaining that Populus is from "an unidentified dveloper". To get around this, follow the instructions at the green arrow; basically, you right-click on the program, tell it to open, and confirm the open request.
In the little pop-up panel, find where you can set the number of generations the simulation runs under "Runtime - Generations", and you will also be able to set the Population Size (N) just below that. On the right panel, we are going to leave "Number of loci" at 6 and "Initial frequency" at 0.5" (Ignore the boxes for "3N Generations" and "Permit selfing?", as well as the options on the right panel.)
Now let's run some simulations with these parameters:
N = 200
Runtime = 100 generations
Number of loci = 6
Initial frequency = 0.5
Click on the green arrows with the word "View" (upper left of the panel, next to the floppy disk, printer, and light bulb icons).
A graph will appear with 6 lines of different colors, one for each of our 6 loci. The frequency of all alleles is at 0.5 when the simulation starts, then the frequencies of each vary independently to simulate genetic drift. Repeat the simulation several times (just keep hitting "View").
If a colored line goes off the bottom of the graph before the last generation, that allele has been lost from the population because of genetic drift. If a line gets to 1.0, it become fixed in the population (it is homozygous) and will remain there. Loss of an allele is a reduction in genetic diversity.
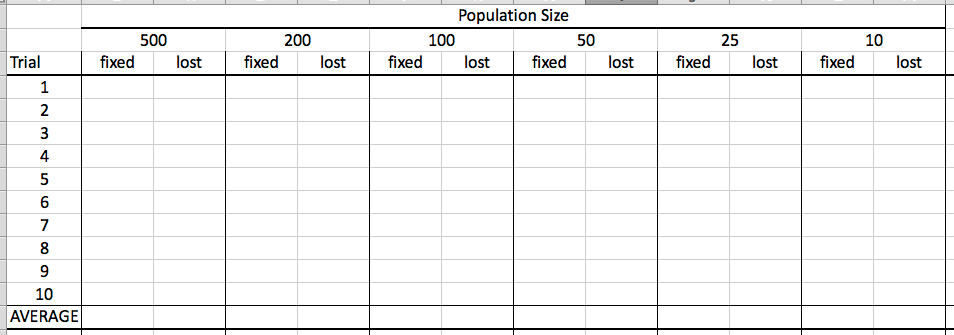
Run the simulation 10 times and for each trial record how many alleles become fixed and how many are lost from the population. Record your data in this table, then repeat with everything the same except the populations sizes, which will be 500, 100, 50, 25, and 10.
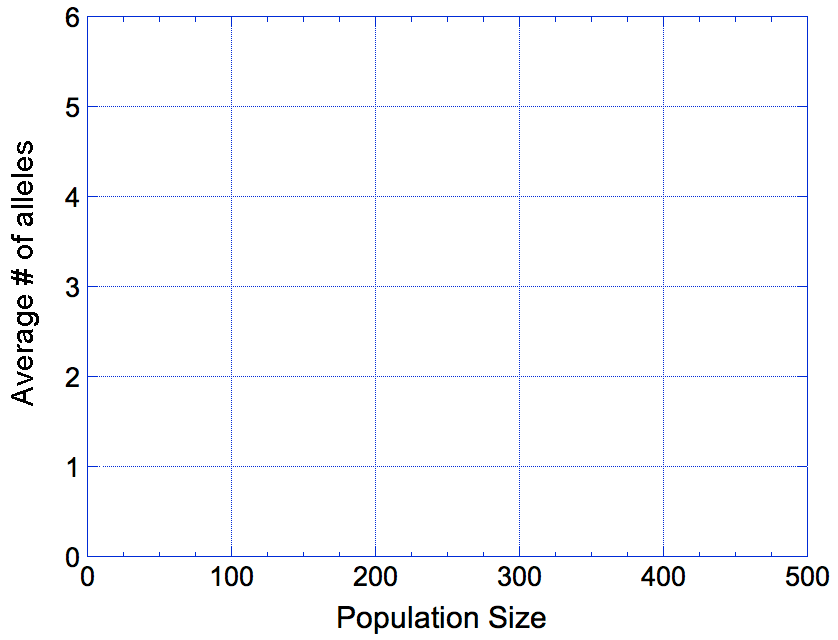
So, looking at the graph you made, you should be able to say something about how the population size of your breed affects the risk of losing a neutral allele just by chance. Think about these questions:
How does population size influence the genetic stability of the gene pool?
If we consider that only the reproductive animals in the population matter for this, what do you think this means for the genetic stability of your own breed?
Has your breed gone through fluctuations in population size (population explosions or bottlenecks)?
What could you do to reduce the loss of genetic diversity by genetic drift in your breed?
Because there is greater loss of genetic diversity at lower population sizes, how would this affect subpopulations in your own breed - e.g., in bench vs show populations, in different countries, different kennels, etc? How would this affect how similar they are to each other? Can you think of a way you could make use of the phenomenon of genetic drift to improve your breeding strategy?
If you would like to learn more about genetic drift, I recommend you have a look at Jeffrey Bragg's article that I mentioned at the top.
*** Population Genetics for Dog Breeders ***
Next class starts 30 March
VISIT OUR FACEBOOK GROUPS
ICB Institute of Canine Biology
...the latest canine news and research
ICB Breeding for the Future
...the science of animal breeding