Most of the traits you care about in a dog (e.g., hunting ability, temperament, intelligence, athleticism, herding prowess) are determined by many genes - hundreds, and maybe even thousands. The process of breeding and selection basically shuffles the genes from the parents, then distributes a different set to each puppy. If a pair of dogs could produce many thousands of puppies, each will be genetically unique because the many possible combinations of alleles create the possibility of huge variation in the expressed phenotypes of a trait.
| If you picked a trait of interest and evaluated it in a large litter of puppies, you could then use the data to generate the "class curve" for that trait in the same way that grades on a biology test might be evaluated. Graded "on the curve", the best animals will be on the tail at the right end, and the worst ones in the tail at the left. Most of the animals will fall somewhere in the middle. This is the so-called "bell curve". It's not the perfect fit for every kind of trait; for some the curve might be asymmetric, with more animals pushed away from the middle and towards one end, or with an exceptionally long tail at one end. But the principles will be the same for what we are talking about here. |
When you think about it, this is how dogs are evaluated at dog shows. How well you do will depend on who else is in the ring with you. If it's ten excellent dogs, and you are ranked #8 among this elite group, from your point of view you will have been "dumped" by that judge. And we've all seen the best of a bad lot win best of breed, awarding the win of a lifetime to somebody with a mediocre dog. Of course, the problem with this method of evaluating traits (or students) is that it doesn't select for a specific level of quality or performance. You might be the top student in your med school class, but if you went to a mediocre school and weren't competing against top students, your rose will smell just as sweet and your parents be just as proud as for the student at the top of the class at Harvard Med.
Now let's go back to evaluating that large litter of puppies. Each has the same set of parents, so the potential genetic "influence" is exactly the same, but of course every one of them has its own unique mix of paternal and maternal alleles, and every one will be just a bit different from all the others. While you're looking at these puppies trying to make up your mind which are suitable to keep for breeding or show, you're thinking in the back of your mind that you want to breed only "the best to the best". In these puppies, you're looking for the one that got the ideal mix of alleles to produce the dog of your dreams.
As in our classroom of school children, you want to identify the A+ dog, the very best one to keep back. A very experienced breeder will know if none of the pups actually deserves an A+, even the one that is the "pick" of the litter, and decide that none of them are worth breeding. This is the "selection" step of breeding animals, and it is every bit as important as the original decision of sire and dam. This is also the step that can do the most damage to the future quality of the breed's gene pool.
Why is that?
For traits that are determined by a large number of genes, getting an A+ puppy that is out at the tip of the right-hand tail of our bell curve isn't going to happen very often. If there are lots of B-quality puppies but no A+ or even an A-, all of the puppies might go to pet homes. The breeder might have relegated to the dust bin an assortment of alleles that, mixed just a bit differently, could have produced a really exceptional puppy.
It does make sense that you should choose the best stock to breed from, but then we really need to define "best". Should the "best" be only the puppies in the top 99%? Or does it make better sense to consider that the top 10% or even 20% are all nice quality dogs that, with the right mate and a fortuitous toss of the genes, could produce that once in a lifetime dog. After all, it's not a list of "good genes" that makes an animal better, but the mix of genes - some of which might actually be having a negative effect - that collectively determines the quality of an animal. Understanding that the secret to a great dog is getting just the right mix, you can see how reducing the diversity of genes in the gene pool might be robbing you of that possibility in future litters.
In the science of animal breeding, the notion we're talking about here - of the cumulative effect of a mix of genes determining the quality of a trait - is called the "additive genetic effect". Let's see how this might work with a simple example.
We will assume that we have a trait that is determined by the genes at three loci, so there are a total of 6 alleles that can influence the phenotype (A, a, B, b, C, and c). We will also assume that we can assign a value to each of these that reflects the "benefit" or improvement for the trait of interest:
B = 20
C = -11
a = 1
b = -2
c = 3
Based on these values, the "best" dog would have the combination of alleles that achieves the highest score. The dog with the best genome for this trait would be [AA BB cc], with a total value of
If you choose to keep only the very best puppy from a litter, the average quality of your breeding stock will improve just a bit by saving those copies of the best genes. But you will be eliminating many more copies of those desirable genes simply because they weren't packaged in a way that you thought was worth keeping for your breeding program.
In fact, some of the puppies in a good quality litter will have received many of the good alleles and only a few of the bad ones, and if you kept some of these better puppies instead of rejecting all but the very best, the average quality of the gene pool of your breeding stock would be higher in the next generation.
| Instead of selecting only the very best of the best, a better strategy to steadily improve your animals every generation is to increase the frequency of the "good" genes in your gene pool and reduce the frequency of the genes that are not contributing to improvement. The best way to do that is to breed more of the "better" dogs. |
| Instead of trying to cull your way to perfection, you would be taking advantage of the property of additive genetic value - selecting for that combination of alleles that will improve the scores of your dogs in the next generation. Many breeders are taught that you should breed only the very best, and that breeding close to increase homozygosity will fix the "good" alleles. But now you might see that this strategy assumes that the genetics of the traits you care about are very simple - that there is a single or just a few alleles that will produce the best phenotype, and if you can select just for those alleles, you'll end up with the best possible dog. |
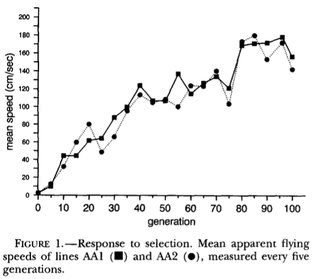
| For example, in a simple demonstration of additive genetic variation, a study selected fruit flies (Drosophila melanogaster) based on their ability to fly upwind in a wind tunnel. Beginning with flies that could fly against a wind of 2 cm/sec, they selected the top 4% of performers every generation. After selection and breeding for 100 generations, they had produced flies that could battle a wind of 170 cm/sec - an astonishing 85-fold improvement in performance (Weber 1996). |
Pushing a trait to such extremes is not necessarily a good idea. Selection for infinite improvement in one trait might reduce the quality of another one. In Holstein cattle, for example, the increase in milk production has resulted in a decline in fertility, which is making it harder to produce more of those terrific milk machines.
But you get the idea - there can be great potential for improvement in a trait if you recognize that achieving it won't come through efforts to narrowly select for a particular "best" gene or genes, but by taking advantage of the additive genetic variation in the gene pool to produce the combinations of alleles that will improve the quality of this generation over the last one.
Are the dogs we have today better than the ones 50 or 100 years ago? They might be. But the one thing that is fairly certain is that the loss of genetic diversity in the gene pool of a breed over the generations has progressively limited the potential additive genetic variation. We have selected strongly for the alleles with large and therefore apparent effects, and not so much for the hundreds of other alleles that could have contributed to improvement, like that little pinch of salt that puts a dish over the top. In whatever ways we have improved dogs, we have at the same time reduced the possibility of producing the truly exceptional dog, the A+ student at Harvard, the one that becomes a legend.
Do the breeders that will come after you a favor. Protect the diversity that is in your gene pool now so it can be remixed to produce better dogs in the future.
*** Visit our Facebook pages ***
ICB Institute of Canine Biology
...the latest canine news and research
ICB Breeding for the Future
...the science of animal breeding