This is also a great example of the amount of useful information that can be extracted from a basic pedigree database. Using tools from population genetics, we can explore the genetic history of the breed from the founders up to the present day. Along the way, we can examine the genetic impact of each of the founders, the consequences of popular sires and bottlenecks, the effects of historical events such as wars, and changes in breed popularity that affected population size. We can see how genetic diversity has been lost over the generations, and determine the current size of the gene pool and the effective ("genetic") population size.
That's a lot of great info and also a good example of how population genetics can help breeders more effectively manage the genetic health of their breed.
Even if the Afghan Hound is not your breed, this should be interesting to you. And if your breed has a pedigree database, it's worth going through this to see some of the amazing things you might be able to learn about the genetic history and present status of your own breed!
- The modern Afghan Hound was developed through selective breeding of land race dogs from Afghanistan.
- Within 20 years, the composition of the gene pool was fixed and has changed little since.
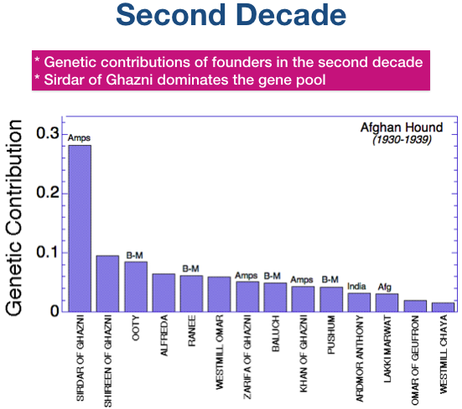
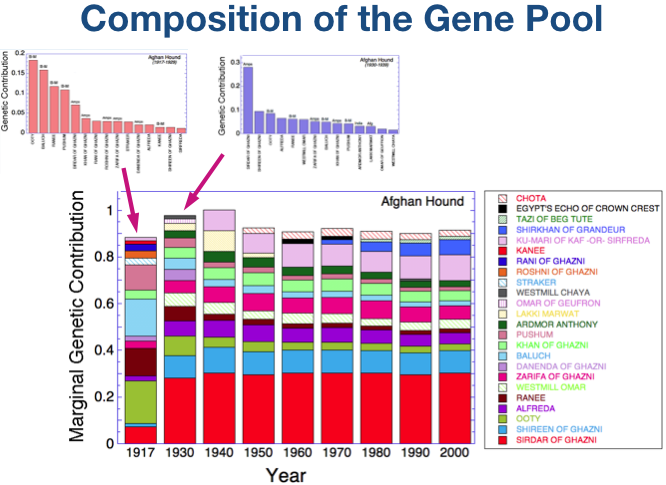
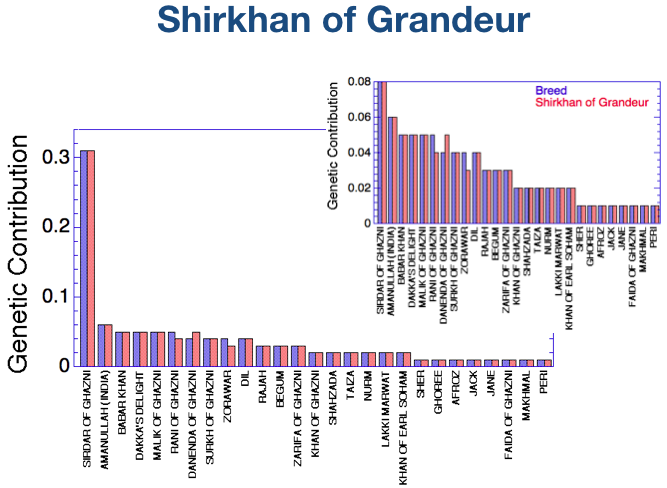
- A single founder dog, Sirdar of Ghanzi, accounts for 30% of the total genetic diversity in the breed today.
- The size of the current gene pool is about 9.
- The average level of inbreeding is roughly 20% from pedigrees and 30% from DNA.
- The average kinship coefficient is less than the inbreeding coefficient, indicating that breeders are preferentially inbreeding.
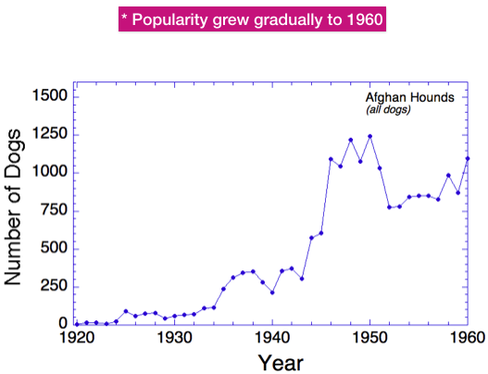
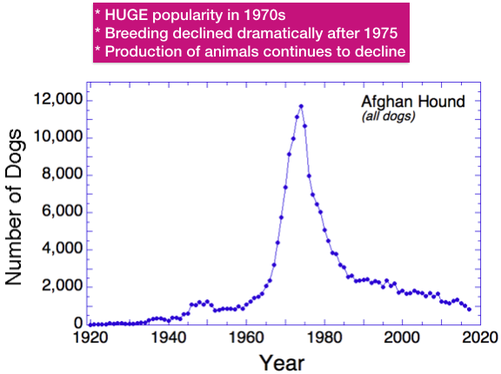
- The number of dogs produced yearly has been dropping for decades.
- The effective population size is falling and is currently at a critical level (about 100).
- Breeders need to protect the remaining genetic diversity and could improve it through introduction of new founders from the country of origin.
- Breeders should reduce multiple breedings and instead try to use multiple puppies from each litter instead of just one.
- Breeders should maintain an up to date pedigree database and do regular analyses like this to stay informed about the genetic status of the breed.
- Breeders should minimize the overall risk of genetic disease through sound genetic management instead of one mutation at a time through DNA tests.
The dogs that were to become the purebred Afghan were brought back from the area by British soldiers and their families. Aggressive selective breeding programs that emphasized elegance, beauty, and coat quality quickly transformed the land race dog into the modern purebred Afghan Hound.
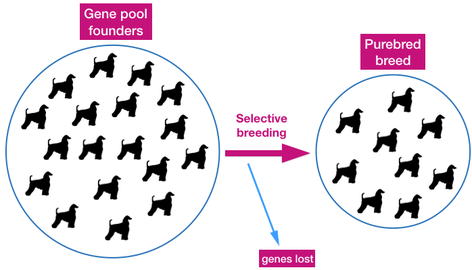
The process of breed development can have a profound effect on the genetic makeup and diversity of the developing breed. Selective breeding refines type to suit the breed standard and the tastes of breeders, and desired traits are locked in using inbreeding.
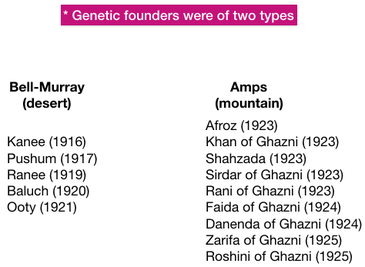
There were two main sources of dogs: short-haired dogs from the steppes that were more suited to the desert, and long-coated dogs from mountainous areas.
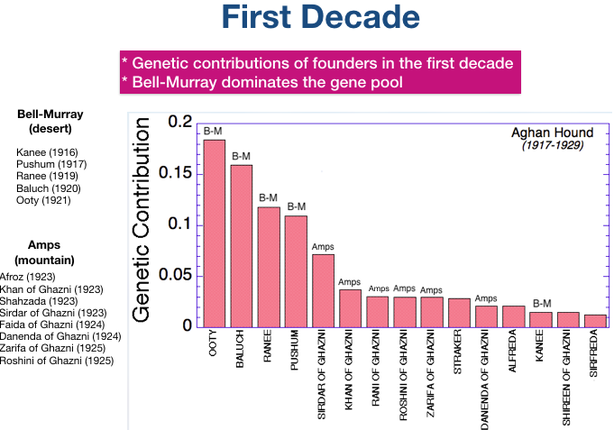
These two populations of founder dogs and the kennels that promoted them are referred to as the Bell-Murray (B-M) dogs (desert type) and the Amps dogs (mountain type).
(Offspring of first generation founder dogs also appear on this chart.)
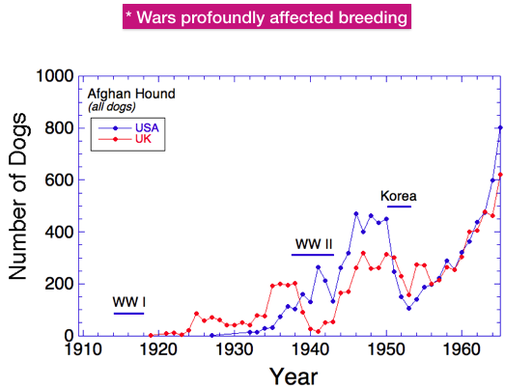
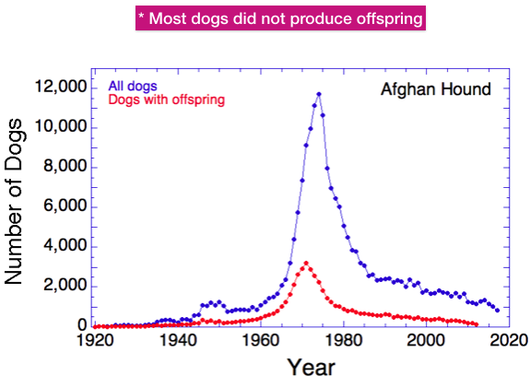
In most purebred breeds, only a fraction of the dogs born produce offspring themselves, usually about 15-40%. This is also true of Afghan Hounds. This graph shows the total number of dogs produced yearly (in blue, as in the previous graph) and also the number born each year that produced offspring (in red). The difference is substantial. The next graph will show the magnitude of that difference.
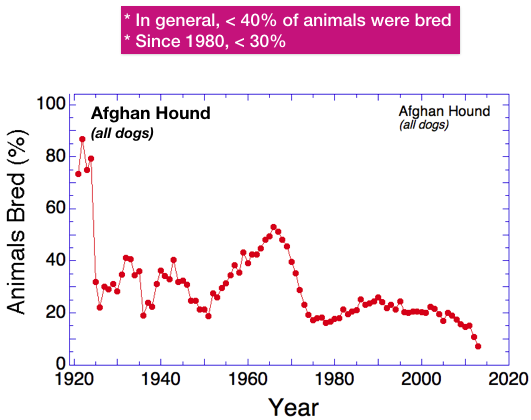
After the early years of breed foundation, the fraction of dogs bred has varied from about 15-50%. There is a peak in 1965 that corresponds to the start of the period of exponential growth. However, while the population was exploding, the fraction of dogs being bred actually declined and didn’t stabilize until about 1975, which corresponds roughly to the peak in the number of dogs produced per year.
Since 1990, the fraction of dogs that produce offspring has been declining from about 25%, and it appears to be dropping rapidly in the last decade. The most recent estimate (for 2013) is only 7%.
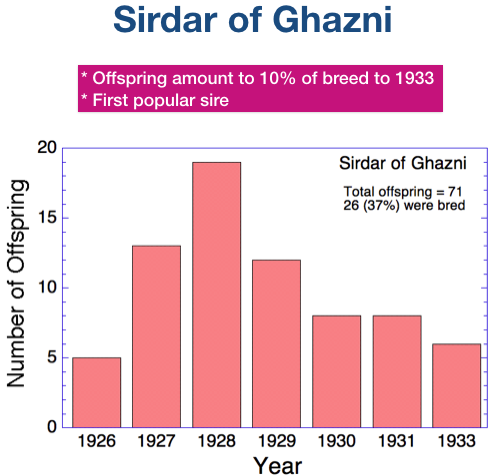
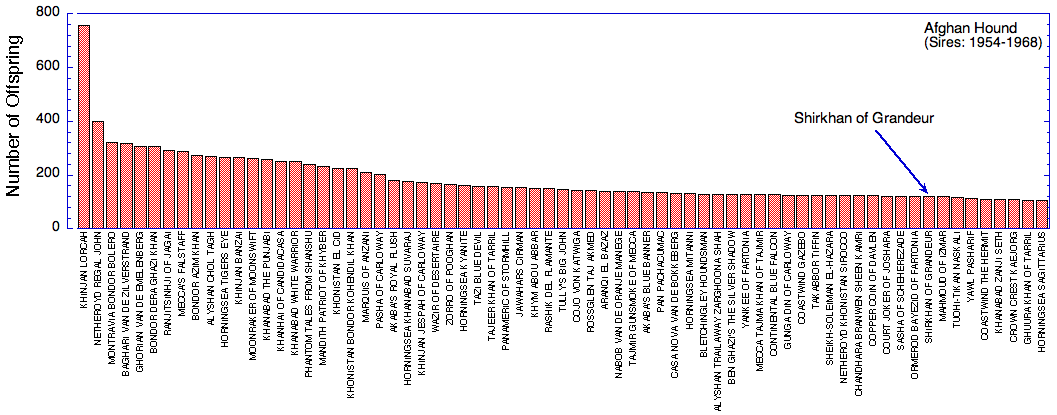
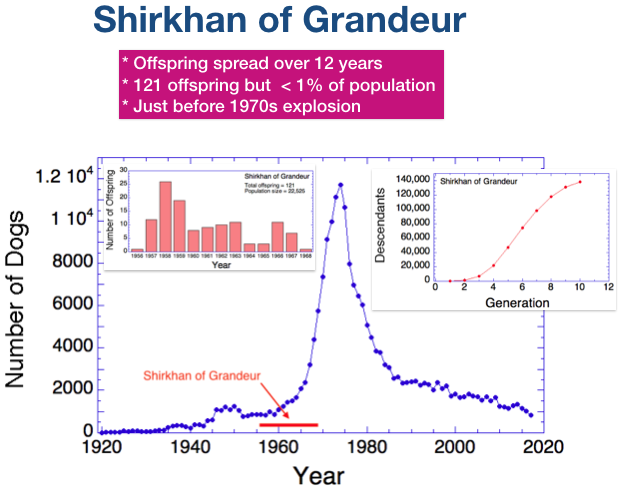
1) His offspring appear just at the beginning of the population explosion that began in the 1960s;
2) MOST of his early descendents were bred.
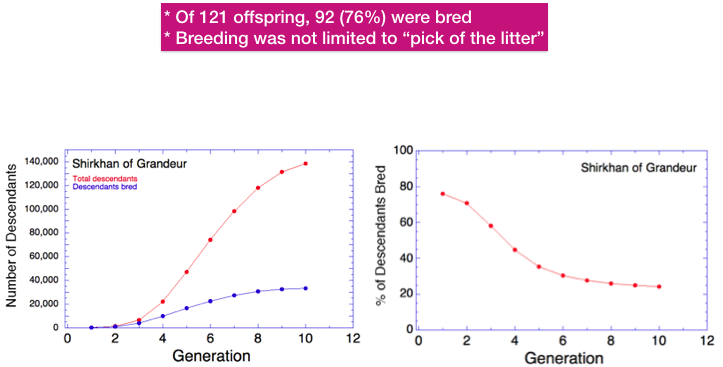
Instead of the usual 20-40% of offspring being bred, for the first 2 generations about 70% of his descendents also produced offspring, and over the next two generations the percentage was more than 40%. His inbreeding coefficient was 8%, so he passed some genetic diversity to his offspring.
The graph on the left shows the number of offspring he produced from 1956 to 1968. The graph on the right is the cumulative number of his descendants over the next dozen generations. After 10 generations, he had > 130,000 descendants. The total number of dogs in the pedigree database between 1955 and 2017 is 197,214.
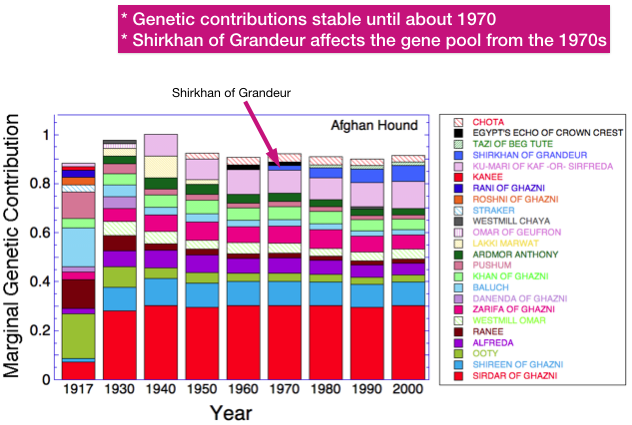
There are just three evident differences - Shirkhan of Grandeur had less genetic contribution from Rani of Ghazni and Zorawar than was present in the population as a whole, and more from Danenda of Ghazni.
Because every dog must have registered parents and all share the same founders, ALL breeding of purebred dogs is to related dogs. This is the definition of “inbreeding”. Breeding related dogs creates homozygosity because related dogs share some of the same genes. Homozygosity refers to the inheritance from an ancestor of the same allele from both parents. There can be close inbreeding (e.g., siblings) or distant inbreeding (third cousins), but it’s all inbreeding - it produces homozygosity.
There are advantages and disadvantages to inbreeding. Inbreeding reduces variation among animals and increases predictability in breeding. It also increases prepotency, so parents produce offspring that share their traits.
But there are also significant disadvantages to inbreeding, mostly related to health. Inbred animals are less fertile and produce fewer offspring, and they are more likely to express diseases caused by recessive mutations.
Because the disadvantages of inbreeding affect the success of your breeding program, we tend to worry a lot about inbreeding.
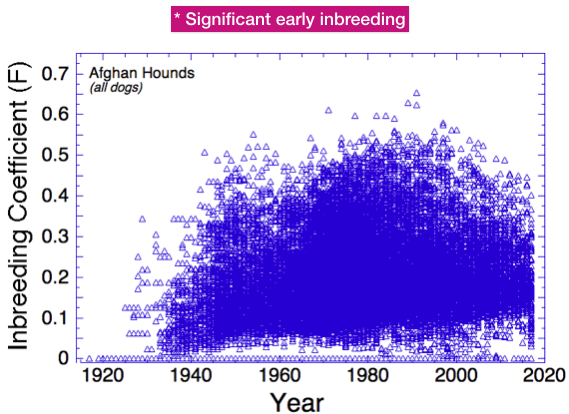
This chart contains the inbreeding coefficient of every dog in the pedigree database for which one can be calculated. The very low or zero values that you see represent dogs with missing ancestors or parents; these values will be underestimates of actual inbreeding so they are an artifact of the data.
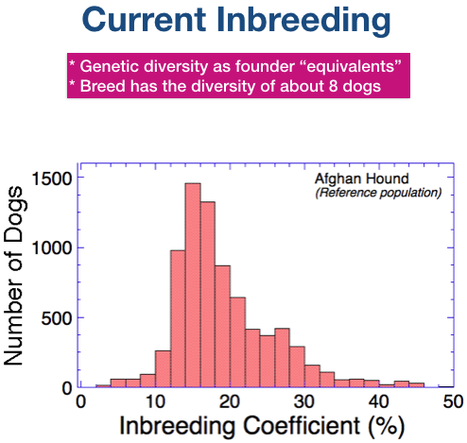
You can see that over the history of the breed, most dogs have inbreeding coefficients higher than 0.05, or 5%. Most dogs fall in the range of 10-30% (0.1 to 0.3).
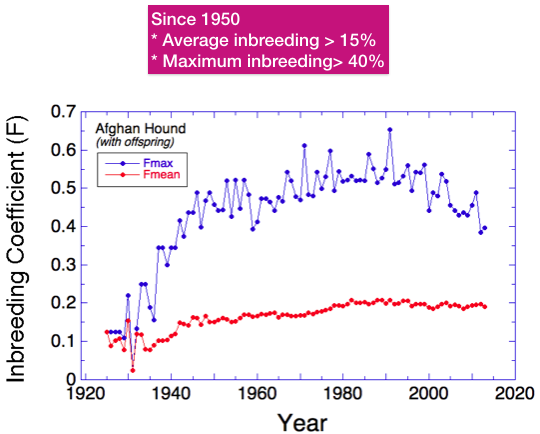
Since about 1935, the average level of inbreeding has gradually increased from about 8% to the current level of about 20%. This is what you would expect for a population with a closed gene pool.
There have also been dogs produced that had extraordinarily high levels of inbreeding. Since about 1950, inbreeding higher than 40% has been common. Since about 2000, the average inbreeding has been stable and the maximum values dropping (but still very high).
What do these values for inbreeding mean? What value would be considered “high”?
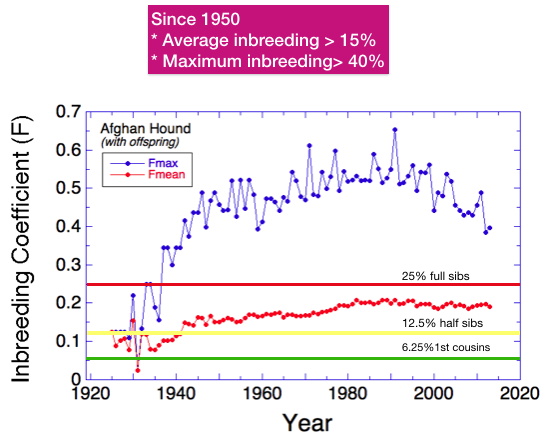
The lines are for inbreeding that would result from crossing full sibs (25%; red), half sibs (12.5%; yellow), and first cousins (6.25%; green). (These assume the parents in each case are not inbred; otherwise these values will be higher.)
Most dogs have an inbreeding coefficient of less than 25%, but there are many higher than 25% and even a few over 40%. Remember that these data are based on analysis of a pedigree database, and we know that missing data will result in underestimation of actual of inbreeding. So these data suggest that inbreeding in general is high in the breed.
Note: The pedigree database I used is not complete. Recent dogs are less likely to be entered, and I looked for the top 40 dogs in the US right now and found only 2, so some countries might less complete than others.
The kinship coefficient (K) is an index of the relatedness (or genetic similarity) of two animals. It is always a comparison of two individuals. The kinship coefficient is related to the inbreeding coefficient: the inbreeding coefficient of a dog is equal to the kinship coefficient of its parents. Similarly, the kinship coefficient of two dogs is the average inbreeding coefficient of their offspring.
We can use the average (mean) kinship coefficient of the animals in a population (mK) to assess how closely related they are to each other. If the mean kinship coefficient is high, then the dogs are, on average, very similar to each other genetically; if the mean kinship coefficient is low, then on average the dogs are less similar genetically.
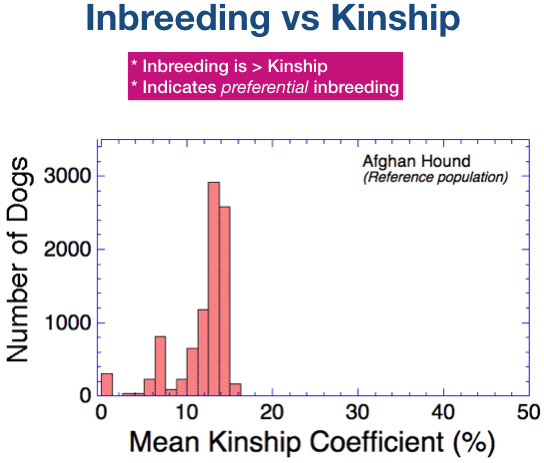
The graph shows the distribution of mean kinship values that would result if the current population of Afghan Hounds was breeding randomly.
(Remember that just as for the inbreeding coefficient, missing data will underestimate this value.)
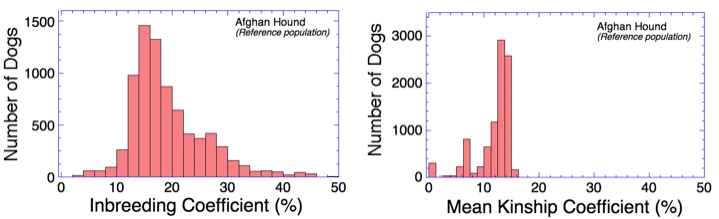
We can learn something about the breeding strategies being used in a population by comparing the data in these two graphs.
Looking at the graph on the right, we can see that if Afghan hounds were breeding randomly, mean kinship coefficients would be relatively low, ranging from 10% to 15%.
From the graph on the left for inbreeding coefficient, we can see that while many dogs fall in the range of 10-15%, most are higher than this.
This also means that with proper mate selection, breeders could reduce the level of inbreeding in their litters by choosing pairs with a lower kinship coefficient.
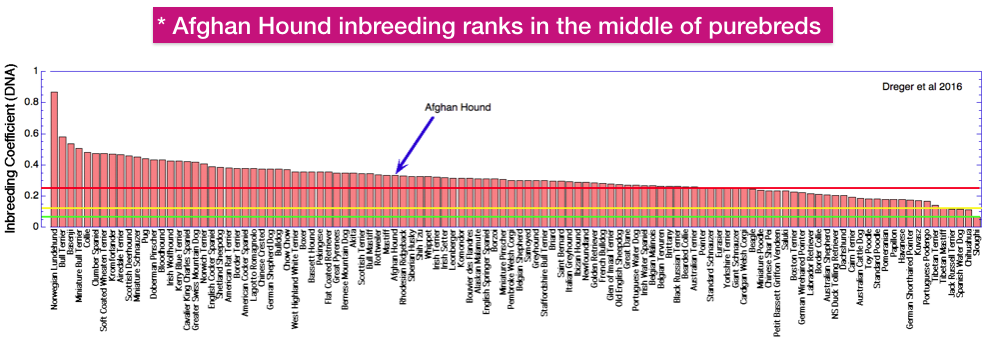
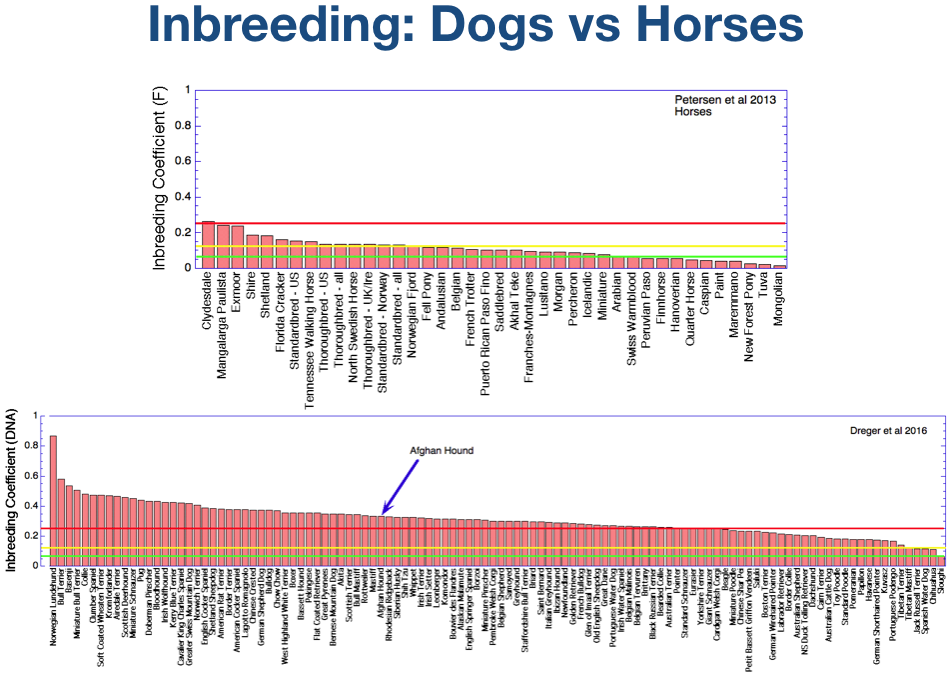
This chart shows the average inbreeding coefficient of a large sample of dog breeds based on DNA data instead of pedigrees. The breed names are probably too small to read, but the Afghan Hound is indicated by the arrow. The inbreeding levels are marked by horizontal lines as before, with red (25%) for a full sib cross, yellow (12.5%) for a half-sib cross, and green (6.25%) for a cross of first cousins.
The average inbreeding coefficient of the Afghan Hound estimated from DNA analysis is about 30%. This is about the “middle of the pack” for the breeds that have been studied.
This compares to about 20% based on pedigree data. Remember, however, that incomplete pedigree data will underestimate actual inbreeding.
The graph at the bottom is the one you just saw for dogs, with Afghan Hound marked with the arrow. The chart at the top is average inbreeding based on DNA for horse breeds, with the same colored lines to mark levels of inbreeding.
Only one breed, the Clydesdale, has an average inbreeding > 25%, and more than half of these breeds are less than 12.5%. In fact, about a dozen breeds average about 6% or less.
Inbreeding in purebred dogs is far higher than in horses, and indeed higher than is typical of other purebred domestic animals. Certainly it is the case that high inbreeding levels are not a requirement of either domestication or of maintaining the traits that make each breed unique.
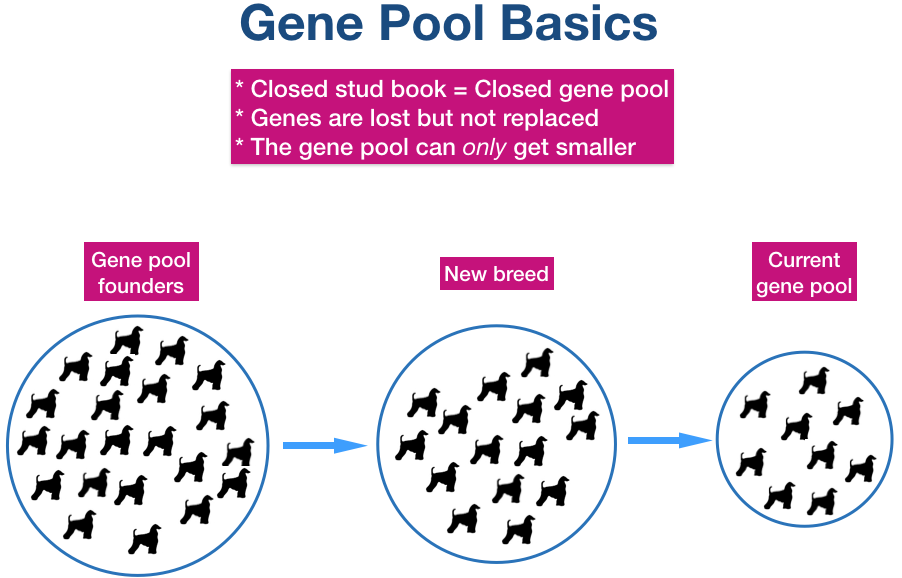
| The founder dogs of a breed, both as original breeding stock and also unrelated dogs introduced into the population later, determine the size of the gene pool. Except for genes added to the gene pool by dogs introduced later, the size of the gene pool will decline over time. All of the genetic variation the breed will ever have is present in these founder dogs. Genetic diversity is lost during breed formation due to selective breeding for development of type and concentration of the genes for those features. Dogs in the current population retain the genes for type, but will have lost genes for other traits and functions over the generations. |
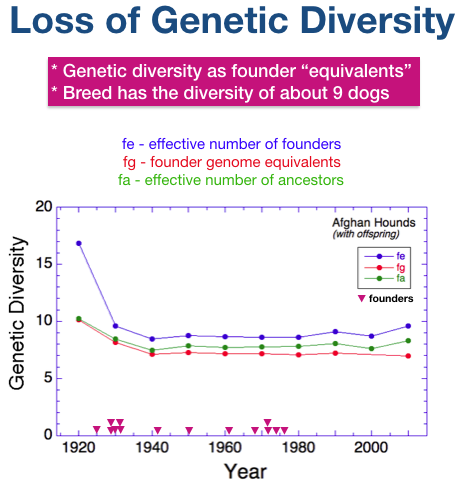
Just look at the blue line for now. This is the line for the “effective number of founders” (fe) - specifically, the number of unrelated dogs that would contain the same amount of genetic diversity as the actual population at a particular time. The highest value, fe = 16.5, occured while the breed is being founded.
By 1930, genetic diversity had declined to fa = 9.5. In another decade, fa is reduced and after that it averages about 9.
Additional unrelated dogs occasionally introduced through the 1970s prevented a reduction in the size of the gene pool.
The size of the current gene pool is about 9.
| Selective breeding affects the composition of the gene pool. Genetic diversity can be lost from the gene pool because of selection - some dogs are bred and some are not. Also, some animals have a disproportionate number of offspring, and their genes become over-represented. The best example of this is the “popular sire”. |

Popular sires are also the most significant cause of genetic disorders in dogs. Most disorders in dogs are caused by recessive mutations that originate with a popular sire. Here’s how this happens.
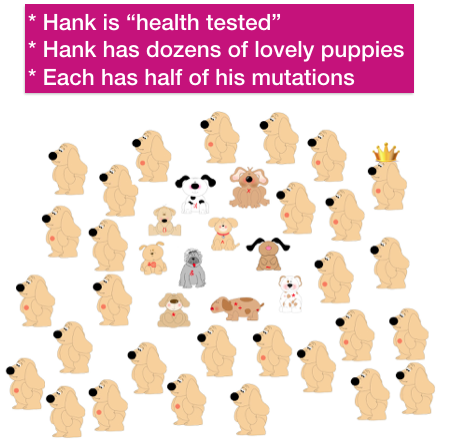
Every dog (every animal, even you!) carries recessive mutations that are harmless if there is only one copy of the defective gene. In this little population of dogs, each has a different mutation (in red) at the arrows. (We’ll pretend each has only one.)
He produces dozens of lovely, healthy puppies. Many other potential sires are not used, and their contribution to the gene pool declines or might be lost completely.
Each puppy inherits half of its genes from its dam and half from Hank. By random chance, half of the puppies produced by Hank will carry one copy of his recessive mutation. They are not affected by it, because they inherited a normal copy of the gene from their dam.
Everyone is thrilled with the stunning, healthy puppies sired by Hank.
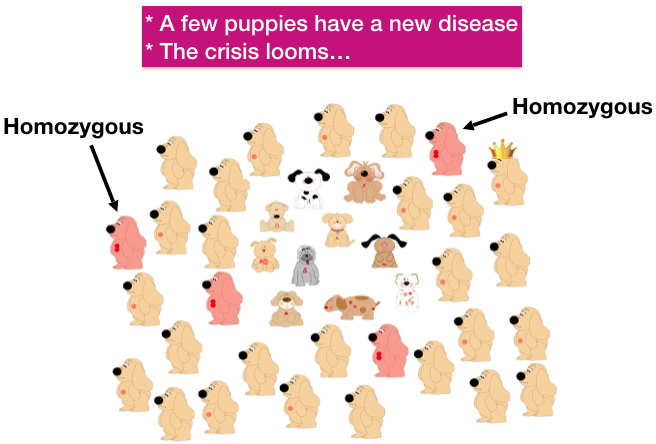
| A generation later, some people have bred half siblings that share Hank as a sire. Some of those puppies inherited a copy of Hank’s mutation from both of their carrier parents, making them homozygous for the bad gene. In this generation, a few puppies are produced with a genetic disorder, but not enough to sound an alarm. |
| In the next generation, however, Hanks mutation has now spread widely through the breed population, and there are many puppies suffering from this new genetic disorder. After reviewing their pedigrees, breeders trace the source of the mutation to Hank. Hank is villified, his owner shunned, and people start breeding around the disorder by avoiding his descendants. Thousands of dollars are invested in a research study, the mutation is identified, and a new test is added to the list of “health tests” every dog is expected to have before breeding. |
Of course, this isn’t Hank’s fault. Genetic disorders from recessive mutations result when rare mutations become common in a population. Popular sires produce many copies of their mutations and the result will be genetic disorders for the descendants that get two copies.
The next big winner will probably also become the next popular sire, and his unique mutation will be passed to dozens or even hundreds of puppies. The cycle will continue, and a new genetic disorder will probably be produced.
We can completely prevent genetic disorders caused by recessive mutations. Don’t create popular sires.
The other way diversity is lost from the gene pool is “just by chance” - called genetic drift.
Because genes are inherited randomly, not every gene in every parent will be copied into a puppy of the next generation. If an allele is rare in the breed, it can be lost entirely. The change in the composition of the gene pool from generation to generation just due to chance is called “genetic drift”.
Most breeders don’t worry at all about genetic drift. If breed populations were very large, the chance that a rare gene would be lost is low. However, if the population size is small, the loss of rare genes through genetic drift can be high.
For population size, it is not the total number of animals in the breed that matter, but just the ones that are breeding. As we have seen, the breeding population is usually only about 20% of the census population. And if those breeding animals are related, the “genetic size” of the population will be even smaller.
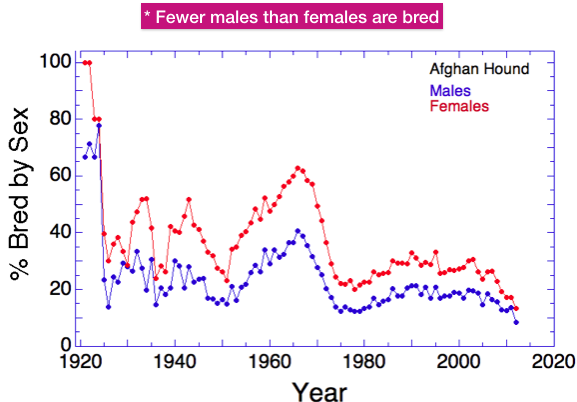
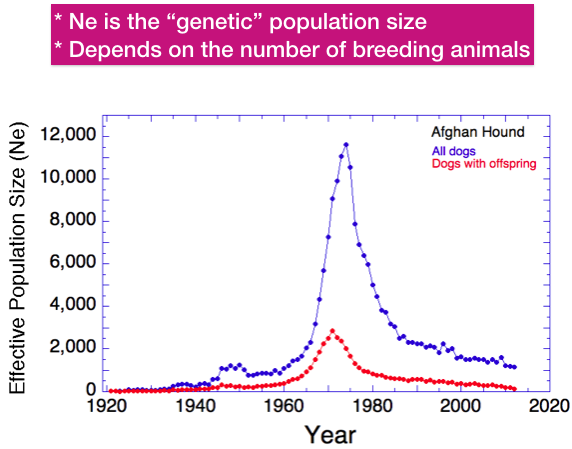
Ne is the size of a hypothetical population that loses genetic diversity at the same rate as the real one of interest. There are many things that can affect Ne, but two important ones are 1) the number of animals bred and 2) the ratio of females to males, both of which we have discussed for Afghan Hounds.
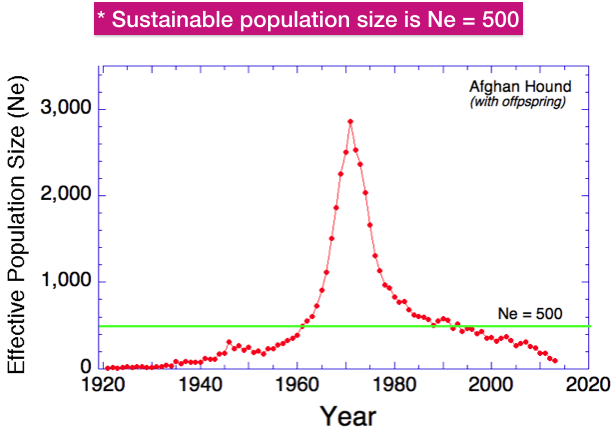
This graph is very similar to one you’ve seen before. But instead of actual number of animals on the y-axis, this one has the effective population size.
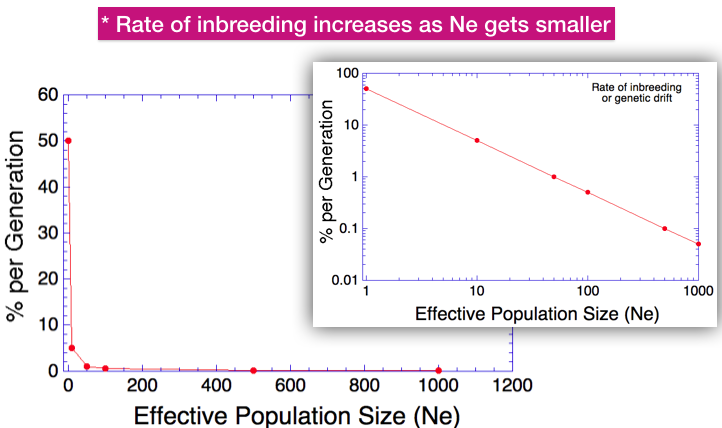
The larger graph (which looks empty) shows how inbreeding (as % per generation) is affected by Ne on linear axes. You can see that for Ne greater than about 100, this line looks nearly flat. Below 100, however, it increases very quickly.
If we log both axes, this curve becomes linear. Now we see in the inset graph that when Ne is small, even a small change will have a large effect on the rate of inbreeding.
What is the message here for Afghan Hounds? As the number of breeding Afghan Hounds continues to decline, we can expect to see inbreeding increase.
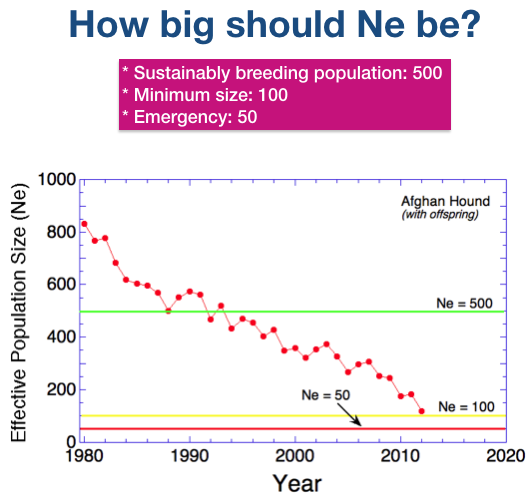
For Ne, how small is too small?
As a general rule of thumb, the effective population size should be about 500. This is considered the size for a sustainably breeding population because the rate of inbreeding will below and there will be low risk of extinction. The effective population size of the Afghan Hound fell below 500 in about 1995 and has declined slowly since.
We are only interested in the most recent decades for Afghan Hounds, so lets change the axes on the graph.
There is a green line at Ne = 500 is as before. A yellow line has been added at Ne = 100, which is considered a minimum population size.
And there is a red line at Ne = 50, which is considered to be the “emergency” value of Ne, below which a population is at risk of extinction from inbreeding depression and genetic disease.
The effective population size of the breed is currently about 100 and will likely continue to decline. Breeders should act to reduce a further declines in Ne by increasing the fraction of dogs used for breeding. You can do this without increasing the number of puppies produced.
A few of these might be caused by single mutations for which a test could be developed - e.g., progressive retinal atrophy, von Willebrand’s. But there are no DNA tests available for the Afghan Hound.
Most of these are polygenic, so genetic management must be through breeding strategy.
Here’s the usual process. A disorder pops up in the breed (popular sire?). We spend money and time to study the disease, look for a gene, and develop a test. For a few problems, we find a gene. For most we do not. And new genetic disorders continue to appear. Dogs are not getting healthier.
We can solve the problem of genetic disorders in dogs faster and better than we are using proper genetic management. Low inbreeding will reduce risk of ALL disorders caused by recessive mutations. Minimize loss of genetic diversity so you don’t lose important genes. (They’re all important!) And avoid popular sires.
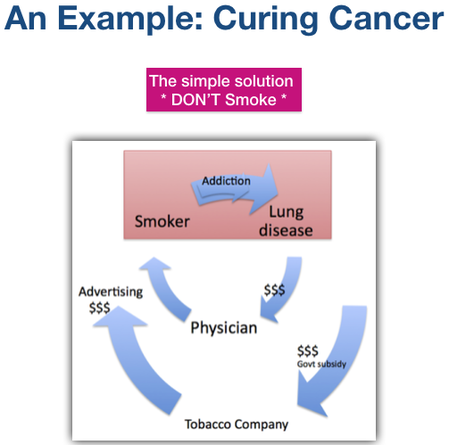
Here’s an analogy -
So we smoked, spent money on cigarettes, doctors, and research, and continued to smoke. This went on for decades, to the delight of the tobacco companies.
Finally, the government launched a war on smoking. There were warnings on cigarette boxes, commercials on TV of people with breathing tubes, and black lungs from autopsies. Smoking was villified. And new methods to help people quit smoking were created - patches, gums, pills, acupuncture, whatever.
People stopped smoking. When was the last time you heard about the scourge of lung cancer? It ceased to be a problem.
The solution was obvious and simple. If smoking causes cancer, don’t smoke.
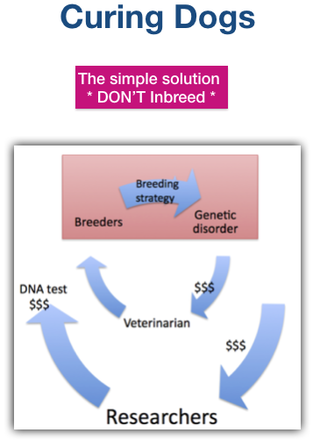
We inbreed deliberately, far more than necessary to produce dogs with consistent type.
Inbreeding adversely affects health in dogs. It increases risk of genetic disease, reduces fertility, shortens lifespan, and much more. We spend a fortune for veterinary care, we spend a fortune on tests and veterinary procedures to produce a litter of puppies and there are fewer of them. We spend money for “health tests” to avoid the genetic problems caused by inbreeding, and we invest a huge amount of money supporting research into the many diseases that have become common in dogs.
Just as for smoking and lung cancer, we feed this cycle. Spending money will never solve this problem. DNA tests will not solve this problem. Research will not solve this problem.
The solution is obvious and simple. Don’t inbreed. Restore the gene pool of your breed, and protect that genetic diversity like money in the bank - because literally, it is. Skip the vet bills, the DNA tests, the donations for research, and all the heartache.
1) Falling popularity and therefore declining population size
2) Inbreeding
3) Small gene pool
4) Popular sires
5) Genetic disorders
Breed smarter. Make the best possible use of the genetic diversity in the gene pool. Avoid multiple breedings, use dogs from underrepresented lines, and breed 2 or 3 pups from a litter instead of just one.
Use information about genetic relatedness (i.e., kinship coefficients) to select less related dogs to breed to. This will reduce inbreeding and the risk of genetic disorders.
NO Popular Sires.
Add fresh genes to the gene pool using dogs from the country of origin. Type can be restored in a generation or two. Health cannot be improved without replacing genes that have been lost. You can’t have type without health.
Keep an up to date pedigree database so you know what animals you have and who they are related to.
Analyze the pedigree data on a regular basis to monitor the genetic status of the breed.
Work with population geneticists to design an efficient strategy for introducing new dogs to the population. Follow the plan so every dog counts.
Use population genetic management instead of mutation tests to reduce genetic disorders.
With sound genetic management and careful breeding by knowledgable breeders, you can continue to produce healthy, beautiful dogs for many generations.
The analyses presented here are based entirely on a pedigree database that was created by me from a database provided to me by the late Jim Coudriet and Peter van Arkel and records in the Afghan Hound International online pedigree database. I am grateful to both of these sources for the diligence in maintaining these valuable records.
I made use of several subsets of data for analyses: “all dogs”, which included all animals in the database; “with offspring” included only dogs with recorded descendents in the database who were born before 2014, since dogs after this have probably not reproduced; and the “current population” or "reference population", which includes all dogs in the database from 2010 to 2018 and are assumed to be potentially reproductive
ICB's online courses
***************************************
Visit our Facebook Groups
ICB Institute of Canine Biology
...the latest canine news and research
ICB Breeding for the Future
...the science of animal breeding