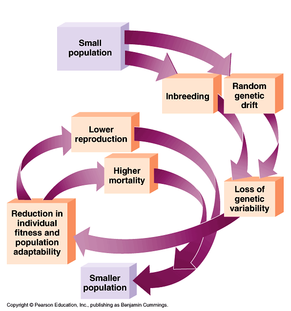
Let's say we have a population of animals that are randomly breeding. Over generations, the average level of inbreeding in the group will increase, just as you can imagine it would if you and a random assortment of unrelated people were shipwrecked on a remote island - eventually, everybody ends up being related to everybody else. This is indeed what happens to populations of animals on islands, and it is also what happens in purebred dogs that have a closed stud book. And the smaller the population, the faster the degree of inbreeding increases.
| We know that inbreeding increases homozygosity, and this can have benefits (e.g., more uniformity, prepotency), but also significant disadvantages that include inbreeding depression (which mostly affects reproductive traits like litter size and puppy mortality, as well as lifespan) and the increased expression of deleterious recessive alleles (more here and here). In wild animals, increasing levels of inbreeding eventually result in a negative feedback loop called the extinction vortex, which we've talked about before. So how do species of wild animals keep from going extinct? First, most avoid inbreeding, and some animals have been shown to be able to detect their level of relatedness (or genetic similarity), possibly from pheromones or the genes of the immune system. (Recent studies are finding this in humans as well.) A preference to breed with animals that are not related is a very effective way to keep inbreeding under control. Another thing they can do is to not keep all their eggs in one basket, so to speak. |
Animal breeders working with relatively small populations of animals (hundreds or perhaps thousands, but not many millions) also need to manage inbreeding or it will increase relentlessly over time. They can do this using breeding strategies that mimic what happens natural populations. How would this work?
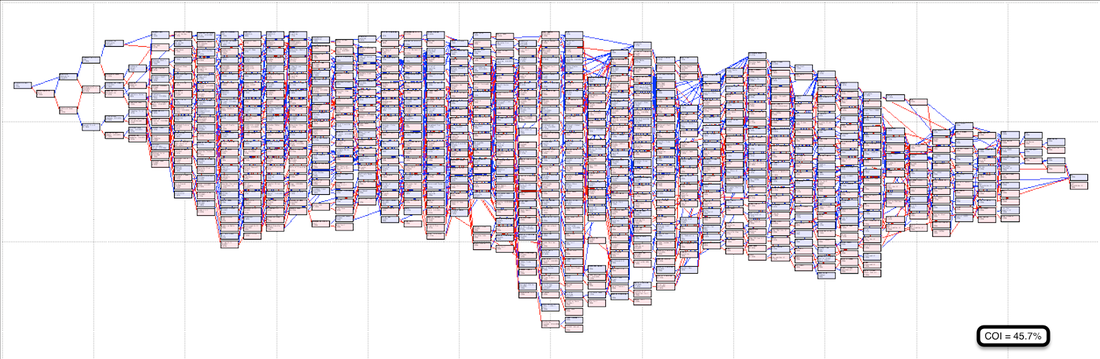
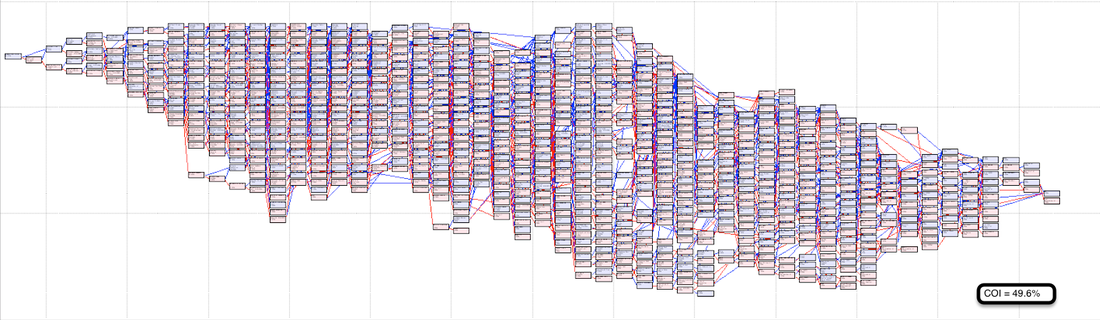
Here are some pedigrees of individual dogs (all the same breed) in which each animal is represented only once (these charts are produced by Pedigree Explorer). Founders are at the base (on the right), and the subject is on the left. (We looked at some pedigrees like this in our discussion of The Problem with Poodles.)
The first two pedigrees are similar and are typical of most of the pedigrees I've looked at of purebred dogs. The most recent generations (on the left) have some tight breeding, and like most breeds there are just a handful of dogs at the base. The coefficient of inbreeding (COI) of the first one is 45.7%; the second is 49.6% (!). A sib-sib mating would have a COI of 25% by comparison. These dogs are definitely inbred.
There are two things to consider.
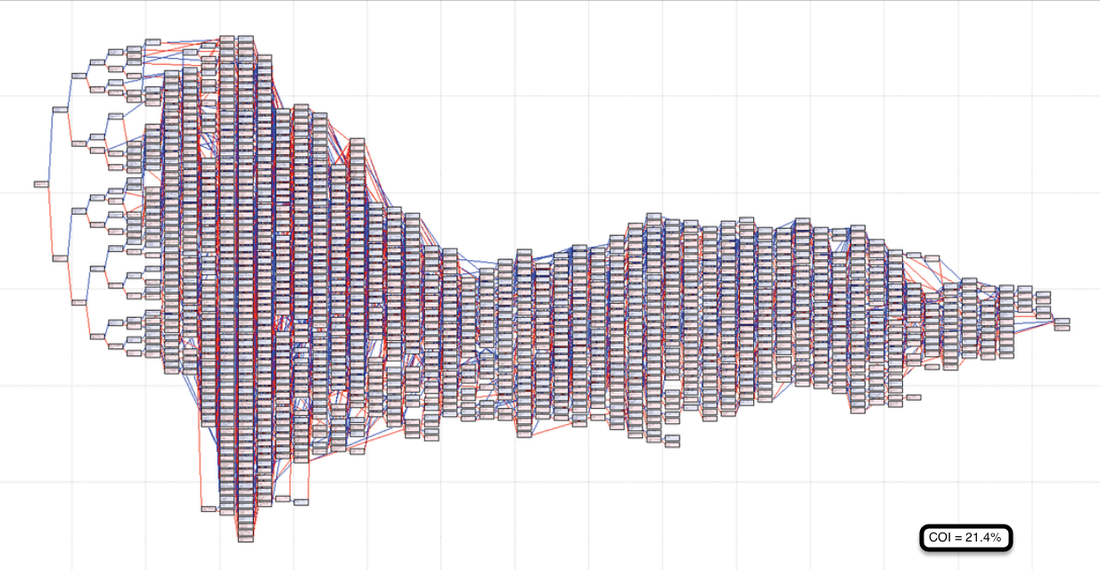
1) The probability of inheriting a particular allele from the pair at a locus of a parent is 1/2, or 50%. With each added generation, that probability is reduced by half; so the probability of inheriting a specific allele from a grandparent is 25%, from a great-grandparent it's 12.5%, and so on. The deeper in a pedigree a particular ancestor occurs, the lower the probability of inheriting a specific allele from that animal. In the bottom two pedigrees above, the parents of the dog share no common ancestors in the most recent generations. In generations, in which there are no shared ancestors, there is no additional inbreeding. So any inbreeding must be deeper in the pedigree, and its effect is reduced by half for each additional generation to the shared ancestor.
2) The other thing we can take advantage of are two forces that will tend to make two populations increasingly different genetically over time - selection, either natural or artificial, and something called genetic drift. You know about selection. If you started with two genetically identical populations, then bred each independently but selected for the same traits in each, you would no longer have identical populations several generations later. In the shuffling of genes with each generation, and the selection of offspring to breed in the next generation, some genes will increase in frequency over time and some will decrease, and some might become fixed and others lost. The offspring in generation 4 of the first population will not be genetically identical to those of the second population, and if you were to breed together an animal from each, their lower genetic similarity will be reflected in a lower coefficient of inbreeding in the offspring.
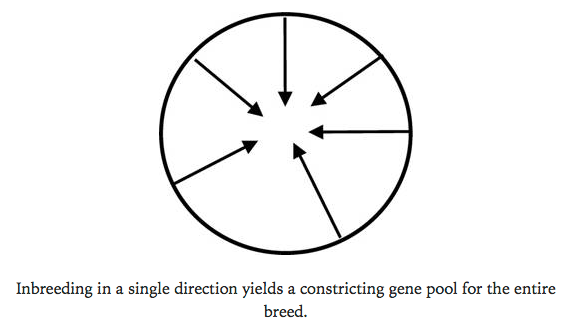
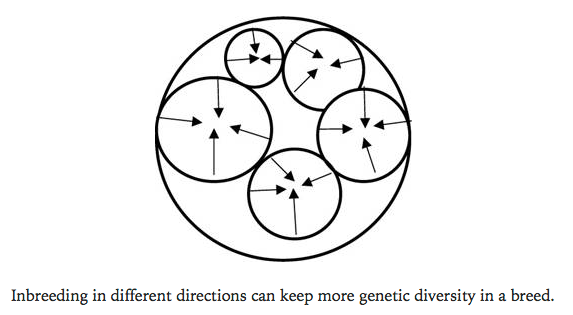
So think about this. You have a breed with thousands of animals in a closed gene pool. If you can divide that population of animals into sub-populations and breed them independently for several generations, you can create for yourself groups of animals that you can use every several generations to produce animals with a lower lower the level of inbreeding, and also reintroduce genes that might be present in one subpopulation but lost from another. In essence, you can use inbreeding in subgroups in a clever scheme to manage inbreeding in the population as a whole.
- Sponenberg DP and DE Bixby 2007 Managing breeds for a secure future: strategies for breeders and breed associations. (Avail as an eBook through iTunes)